Diabetic foot ulcers (DFU) in patients with uncontrolled diabetes mellitus are considered a global public health menace that is highly associated with morbidity and mortality. Pathogenic microorganisms entrenched deep into diabetic foot wounds are the causative agents for delayed healing and escalation of diabetic foot wound severity. Pseudomonas aeruginosa is a common opportunistic pathogen associated with several nosocomial infections, cystic fibrosis, and one of the most critical pathogens often isolated from acute and chronic diabetic foot ulcers. The organism can exhibit resistance to a wide range of antibiotics like ciprofloxacin, cefotaxime, and meropenem, thereby causing severe damage to the host tissues, followed by amputation of the affected foot region. Due to their ability to synthesize biofilms, the wound becomes more chronic and incurable, posing a serious threat to immunocompromised diabetic patients. This review highlights on the insights of pathophysiology and microbiological profile of Diabetic foot ulcers, the resistance mechanisms, and the therapeutics available for dealing with drug-resistant Pseudomonas, which could help clinicians in treating DFUs.
Pseudomonas aeruginosa, Diabetic Foot Ulcers, Biofilm, Antibiotic Resistance, Quorum Sensing
Diabetes mellitus is one of the oldest metabolic diseases known to mankind. It is characterized by hyperglycaemia resulting from low insulin secretion or increased glucagon production or insulin resistance. Diabetes is mainly of two types based on the absence or minimal secretion of insulin and reduced response to insulin to peripheral receptors. Type I Diabetes Mellitus (TIDM) is caused due to autoimmune destruction of β-cells by ICA8 and anti-GAD65 autoantibodies. Exogenous insulin treatment is necessary because TIDM typically manifests in children and adults before the age of 30, when blood insulin levels are lowered and patients stop responding to the anti-diabetic regimen. Type II Diabetes Mellitus (TIIDM) is caused by the development of insulin resistance due to a sedentary lifestyle, comorbidities, and other metabolic disorders.1 According to an epidemiology survey in 2021, globally, approximately 536 million people are suffering from diabetic mellitus, with a total anticipated cost of 966 billion USD for diabetes-related healthcare.2 In India, 77 million are diabetic, which is anticipated to climb to nearly 134 million by 2045.3 Diabetes is associated with many complications and is the primary cause of neurological disease, cardiovascular disease, kidney failure, blindness and lower limb amputation.4,5
Diabetic foot ulcer (DFU), a well-known TIIDM-associated complication, is a primary cause of hospitalisation, accounting for 20% of all hospital admissions and morbidity.6,7 Approximately 58% of patients with foot ulcers are prone to septicaemia.8 Untreated DFU can progress into ulcers and gangrene eventually leading to limb amputation and death.8,9 It has been estimated that DFU is the cause of 50 to 70% of limb amputations.10 In addition to morbidity, DFUs have significant socioeconomic repercussions. The average cost of hospital admission for amputation in the US is around $100,000.11,12 The price of treatment and management varies by nation, from $188,000 in the US to $3060 in Tanzania.13 The price of DFU therapy in India, which has one of the highest rates of diabetes, is roughly $1960.14 In India, it is anticipated that it will take 5.7 years of a patient’s income to treat a DFU.15
A deeper understanding into the systemic progression of the infections, treatment and management of the infection is required. Only few studies are available to understand the correlation of Pseudomonas in diabetic foot ulcers. Hence, the present review aims to discuss the pathogenesis of diabetic foot ulcers emphasizing the role of multidrug resistant Pseudomonas aeruginosa in the progression of diabetic foot infections and the existing therapeutics available to combat the resistance which could be further studied to deal with DFU complications.
Pathophysiology of diabetic foot ulcer
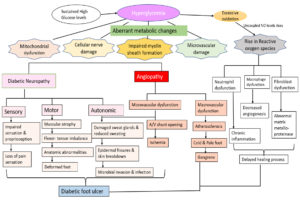
DFU is often manifested by lesions and abrasions in the skin, but its aetiology is multifactorial. The pathophysiology of DFU is attributed to a complex triad of peripheral neuropathy, vascular foot abnormalities, arterial occlusive damage and decreased immune response to infection.16,17 Hyperglycaemia induces aberrant metabolic changes, such as an increase in intracellular glycosylated nerve proteins, protein kinase C activation, increased hexosamine flow, and polyol pathway, all of which lead to nerve injury.18 Studies have shown that motor neuron damage causes an imbalance in flexor-extensor coordination and the development of anatomic abnormalities such as Charcot’s foot, hammerhead toes, and claws.18,19 Damage to sensory nerves results in loss of sensation and proprioception, which lowers the pain threshold, making the foot more susceptible to heat and trauma, thereby increasing the risk of foot ulcers. Autonomic nerve damage inhibits sweat glands, and the foot’s capacity to moisten skin may deteriorate, resulting in epidermal fissures and skin breakdown, providing viable channel for microbial invasion and infections.7,20,21 These neuropathy-related impairments result in “high-pressure” zones at the metatarsal head on the plantar surface of the foot.22 Hyperglycaemia-induced vascular alterations in the peripheral arteries resulted in a decrease in vasodilators and increased plasma thromboxane A2 levels. As a result, peripheral arteries experience vasoconstriction and plasma hypercoagulation, which ultimately increases the risk of ischemia and ulceration.5 Additionally, immunological alterations enhance T lymphocyte apoptosis, which lowers the foot ulcers’ ability to heal.23 Due to these cellular and metabolic changes, diabetic patients experience repetitive trauma from walking in combination with decreased sensation and proprioception. This leads to the dislocation of the protective plantar fat pads, which can result in ulceration and infection from inadequate skin protection or bad footwear.24
Patients’ poor attention to their skin, failure to recognise cutaneous injuries (redness, blister formation), or delayed treatment can result in the progression of foot lesions to ulcers and the development of microbial invasive soft tissue infection. Eventually, the infection penetrates into the deep skin layers and spreads to the midfoot muscles, joints, and tendon sheaths. As the infection progresses, the deep tissue fills with pus, leading to tissue necrosis and abscess formation. One-half of major (above- or below-knee) lower extremity amputations in people with diabetes are due to microbial infection.7,25 Flowchart depicting the pathophysiology of diabetic foot ulcer is given in Figure 1.
Microbiology profile of diabetic foot ulcers focussing the different regions of India
Diabetes foot wounds have a complicated microbiome. A diabetic foot ulcer is caused by recurring infections from aerobic, anaerobic and fungal microorganisms, either singly or in combination.26 Microorganisms isolated from diabetic food wounds were identified using 16S rRNA, short gun metagenomic and pyrosequencing.27,28 According to the American Infectious Disease Society, DFUs are divided into three subcategories: mild infections with superficial symptoms, moderate infections with deeper and pronounced symptoms, and severe infections with systemic symptoms or metabolic abnormalities.29 Mild infections appear to have a simpler microbiota inhabited by common skin commensals such as beta-haemolytic Streptococcus (S. agalactiae; S. pyogenes; S. mitis), aerobic Gram-positive cocci (Staphylococcus aureus), Coagulase negative Staphylococcus epidermidis, which have been identified as pathogens in foot ulcers.27 Among these isolates, S. aureus was reported to be the most common isolate ranging from 25% to 30% from early diabetic wounds.30,31 Group B Streptococcus associated infections are rising in the present days leading to soft tissue damage and severe blistering cellulitis followed by amputation.32 Moderate infection wounds are primarily colonised by aerobic, non-fermenting Gram-negative bacilli i.e., Enterobacteriaceae (Escherichia coli, Morganella morganii, Klebsiella pneumonia, Proteus mirabilis), Pseudomonas aeruginosa, Citrobacter spp., Enterobacter spp. and Acinetobacter spp. In several studies, Gram-negative isolates (50%) were found to be higher than Gram-positive isolates (30%).33,34 P. aeruginosa, which accounts for 14% of the isolates, is a terrible and vital pathogen with antibiotic resistance that can cause significant tissue damage and lead to sepsis and amputation.35,36 Chronic infection wounds are inhabited by aerobic Gram-positive cocci, aerobic Gram-negative bacilli and anaerobic pathogens (Bacteroides fragilis; Propionibacterium spp; Clostridium spp; Peptostreptococcus spp).37 Table 1 summarises the microbiological profile of diabetic foot ulcers done using microbial culture techniques in different geographical locations of India, where 23.7% of DFU cases in North India,38 29.8% in South India,39 11.89% in North-East India,40 11.7% in East India,41 and 27% in Western India42 are caused by Pseudomonas aeruginosa at different study periods.
Table (1):
Microbiology profile of diabetic foot ulcer in different geographical regions of India
Region |
North India |
South India |
North-East India |
East India |
West India |
|---|---|---|---|---|---|
Total number of patients |
102 |
77 |
150 |
148 |
100 |
Study duration |
Dec, 2008 to Feb, 2010 |
May, 2003 to March, 2004 |
Feb, 2015 to Jan, 2016 |
– |
July to October 2012 |
Total Aerobes |
152 |
113 |
182 |
240 |
92 |
Gram-positive aerobes |
55 (36.1%) |
48 (62.3%) |
73 (40.1%) |
88 (36.6%) |
20 (21.73%) |
Staphylococcus aureus |
37 (24.3%) |
19 (24.5%) |
46 (24.86%) |
72 (30%) |
6(7%) |
Enterococcus faecalis |
5 (3.2%) |
3 (3.8%) |
27 (14.59%) |
8 (3.3%) |
|
Beta hemolytic streptococcus |
5 (3.2%) |
– |
– |
4 (1.7%) |
|
CONS |
4 (2.6%) |
20 (25.9%) |
– |
4 (1.7%) |
2 (2%) |
Coryneform sp. |
4 (2.6%) |
– |
– |
– |
– |
Corynebacterium jeikeium |
– |
3 (3.8%) |
– |
– |
– |
Bacillus subtilis |
– |
3 (3.8%) |
– |
– |
– |
Gram-negative aerobes |
97(63.8%) |
65 (84.4%) |
109 (59.8%) |
152 (63.3%) |
72 (78.26%) |
Escherichia coli |
41 (42.2%) |
17 (22.0%) |
37 (20.0%) |
26 (10.8%) |
15 (17%) |
Pseudomonas aeruginosa |
23 (23.7%) |
23 (29.8%) |
22 (11.89%) |
28 (11.7%) |
25 (27%) |
Klebsiella oxytoca |
11 (11.3%) |
1 (1.2%) |
– |
2 (0.8%) |
– |
Klebsiella pneumoniae |
9 (9.2%) |
9 (11.6%) |
22 (11.89%) |
22 (9.2%) |
– |
Klebsiella sp. |
20 (22%) |
||||
Proteus vulgaris |
5 (5.1%) |
1 (1.2%) |
– |
16 (6.7%) |
– |
Proteus mirabilis |
2 (2.0%) |
8 (10.3%) |
9 (4.86%) |
10 (4.2%) |
– |
Proteus sp. |
4 (3%) |
||||
Acinetobacter spp. |
5 (5.1%) |
– |
7 (3.78%) |
12 (5.0%) |
2 (2%) |
Morganella morganii |
1 (1.0%) |
– |
4 (2.16%) |
– |
– |
Citrobacter koseri |
– |
2 (1.2%) |
– |
2 (0.8%) |
– |
Citrobacter freundii |
– |
1 (1.2%) |
1 (0.54%) |
10 (4.2%) |
– |
Klebsiella ozaenae |
– |
1 (1.2%) |
– |
– |
– |
Enterobacter aerogenes |
– |
1 (1.2%) |
6 (3.24%) |
22 (9.2%) |
– |
Edwardsiella tarda |
– |
1 (1.2%) |
– |
– |
– |
Serratia spp. |
1 (0.54%) |
– |
– |
||
Stentrophomonas maltophilia |
2 (0.8%) |
– |
|||
Total Anaerobes |
17 |
5 |
– |
21 |
– |
Gram-positive anaerobes |
15 (88.2%) |
2 (2.5%) |
– |
7 (33.3%) |
– |
Peptostreptococcus sp. |
6 (35.2%) |
2 (2.5%) |
– |
7 (33.3%) |
– |
Peptostreptococcus anaerobius |
4 (23.5%) |
– |
– |
– |
– |
Propionibacterium sp. |
3 (17.6%) |
– |
– |
– |
– |
Clostridium perfringens |
1 (5.8%) |
– |
– |
– |
– |
Eggerthella lenta |
1 (5.8%) |
– |
– |
– |
– |
Gram-negative anerobes |
2 (11.7%) |
3 (3.8%) |
– |
14 (66.7%) |
– |
Bacteroides ureolyticus |
2 (11.7%) |
– |
– |
14 (66.7%) |
– |
Bacteroides fragilis |
3 (3.8%) |
– |
– |
– |
|
Reference |
[38] |
[39] |
[40] |
[41] |
[42] |
Polymicrobial colonization in diabetic foot Ulcers
Chronic wound infections are typically polymicrobial with varied aerobic Gram-negative bacilli and obligate anaerobic bacteria. According to reported studies, polymicrobial infections were estimated to occur in 75% to 83% of chronic DFU cases.34,43 Polymicrobial infections could be induced by anaerobes interacting with aerobes, such as the interaction of E. coli with B. fragilis. As aerobic bacteria multiply, they use oxygen and enhance the growth conditions for anaerobic bacteria, assisting anaerobes in dealing with the harmful effects of oxygen. Furthermore, microbial isolates from wounds were found to tolerate and thrive at a wider pH range, which helps them circumvent the limitations of the external macroenvironment and promote the growth and survival of microbial communities. As a result, the production of virulence factors such as hemolysins, collagenases, proteinases, and short-chain fatty acids gets increased, which promotes inflammation and hinders the healing of wounds.6,44-46 An additional pathogenic property of many organisms is their ability to become enveloped in biofilm. Due to hyperglycaemic condition, P. aeruginosa or S. aureus synthesizes thick biofilms, decreasing antibiotic susceptibility and hindering wound healing.47 These biofilms serve as barriers that inhibit the diffusion of antibiotics, antimicrobial proteins, lysozymes, and defensins, while simultaneously protecting organisms from phagocytosis and promoting antibiotic resistance.48
The growing rate of isolation of antibiotic-resistant pathogens, particularly methicillin-resistant S. aureus (MRSA), glycopeptide-intermediate S. aureus (GISA), vancomycin-resistant enterococci (VRE) and highly resistant P. aeruginosa strains has become a significant problem. Many organisms’ capacity to form biofilm encapsulations is another trait that makes them harmful.29,49 Also, recent studies showed there was an increased incidence of P. aeruginosa in diabetic wounds, especially in geographical locations with hot and humid climates and its management is highly challenging.50 Understanding the physiology involved in making the organism highly resistant to antibiotics is critical. The following sections review the role of Pseudomonas biofilm production, antibiotic resistance, and treatment options available in managing infections.
Antibiotic resistance mechanism in Pseudomonas aeruginosa
Diabetic foot ulcers is a very serious complication of diabetes mellitus which exhibits polymicrobial colonisation and treatment of DFU caused by Pseudomonas aeruginosa is extremely challenging due to its propensity for antibiotic resistance. The patients with DFUs are generally treated using empiric antibiotic therapy and antibiotics such as ceftazidime, cefepime, piperacillin-tazobactam, imipenem, or meropenem are commonly used in these scenarios.51 The organism developed several mechanisms of antibiotic resistance: production of beta-lactamase enzyme for drug inactivation, restrictive outer membrane uptake and efflux mechanism, mutational changes of targeted enzymes or proteins and formation of biofilms.52
Production of beta-lactamase and aminoglycosides modifying enzymes
P. aeruginosa possesses two genes- the inducible ampC gene and the regulatory gene ampR. Mutations in ampR gene trigger the ampC gene’s overexpression and the production of beta-lactamase protein. Beta-lactamase can hydrolyze the amide bond in the beta-lactam ring, thereby inactivating beta-lactam antibiotics.53 Downregulation of ampR was found to be due to excessive ceftazidime treatment.54 Based on Amino acid sequences, beta-lactamases are further classified into A, B, C and D. Class A, C and D have serine Amino acid residue in their active site and were reported to hydrolyze beta-lactam ring. Class B, also known as metallo-lactamases, requires Zn2+ ions for hydrolysis of the beta-lactam ring. Some Pseudomonas isolates were reported to synthesize another type of beta-lactamases known as extended-spectrum beta-lactamases (ESBLs) that has the ability to hydrolyze beta-lactam ring of majority antibiotics such as cephalosporins, penicillin and aztreonam.55-57
P. aeruginosa is also highly resistant to aminoglycosides antibiotics, which contain an aminocyclitol ring linked to amino sugars by glycosidic bonds. P. aeruginosa modifying enzymes- aminoglycoside acetyltransferase, aminoglycoside phospho-transfer and aminoglycoside nucleotide transferase catalyse the structural modifications and inactivation of kanamycin, streptomycin and neomycin.58,59
Outer membrane barrier and efflux mechanism
The outer membrane of Pseudomonas is highly selective and made of specific porins (OprB, OprD, OprE, OprO, OprP), non-specific porins (OprF) and efflux porins (OprM, OprN and OprJ).59 OprF was found to be a major porin for the transport of ions and carbohydrates but has low permeability to antibiotics.60 The OprD is the main porin for the influx of antibiotics, specifically charged lysine molecules. It contains a binding site for carbapenems and its absence in the bacterial cell causes carbapenem resistance. Aminoglycosides, antibiotics and colistin only cross the membrane via binding to lipopolysaccharides outside the cell membrane. Studies on laboratory Pseudomonas strains showed that overexpression of OprH (gated porin) blocks lipopolysaccharides, thereby preventing the influx of antibiotics through the membrane.61
In general, bacterial efflux pumps are mainly used for the extrusion of toxic elements. In P. aeruginosa, four main efflux pumps are used for expelling antibiotics.62 MexAB-OprM is able to pump out beta-lactam antibiotics and quinolones,63 MexXY-OprM extrusion of aminoglycosides,59 MexEF-OprN expels mainly quinolones,64 and MexCD-OprJ expels only β-lactams.65 Overexpression of these genes were reported to increase antibiotic resistance contributing to the development of resistance to multiple drugs.66
Mutational changes of targeted enzymes or proteins
Pseudomonas aeruginosa poses a significant challenge in clinical settings due to its ability to undergo mutational changes within the genome thereby preventing the binding of specific antibiotics. For example, mutations in the mutations in the gyrA and parC genes, encoding for bacterial DNA gyrase and topoisomerase IV respectively, prevent the binding of fluroquinolones.67,68 Similarly, mutations in the genes encoding for Penicillin binding proteins (PBPs) confer beta-lactam resistance. Another mechanism involves mutations that reduce the bacterial cell wall permeability towards antibiotics, viz., aminoglycosides and carbapenems, altering the porin channels such as oprD. On the other hand, mutations in mexR, nalC and nalD responsible for the regulation of the mexAB-oprM efflux pump system, results in its overexpression, causing resistance towards antibiotics along with biofilm formation.69 These mutational changes in the bacterium plays a crucial role for its survival under stress and hence understanding the underlying mechanisms would be critical in developing strategies towards combatting antibiotic resistance.
Pseudomonas biofilm
Biofilm is a matrix of extracellular polymeric substance (EPS) embedded with aggregate microbial communities and helps in the colonization and attachment of microbial cells to the surface. It safeguards the colonized organisms from fluctuating environmental conditions and prevents antibiotic entry, thereby decreasing antibiotic susceptibility. Pseudomonas aeruginosa is well-known for its biofilm synthesis, making it an ideal model for studying biofilm development.
The main components of the P. aeruginosa biofilm matrix are polysaccharides, extracellular DNA (eDNA), proteins, and lipids. The three main exopolysaccharides, Psl, Pel, and alginate, play a major role in the biofilm initiation, attachment of organisms to the surface and maintaining the stability of the biofilm.70,71 Pel (pellicle), a cationic glucose-rich polysaccharide and Psl (polysaccharide synthesis locus), a neutral mannose-rich pentapolymer comprising mainly mannose, glucose and rhamnose, are present in non-mucoid strains of P. aeruginosa whereas alginate present in only mucoid strains.72,73 The primary structural elements of the matrix, Psl and Pel, are important for developing biofilms in early stages, sessile cell adhesion to surfaces, improving cell-to-cell attachment i.e., aggregate formation, and maintaining the structural stability of the biofilm architecture. Additionally, Psl functions as a signalling molecule to encourage increased cyclic 3′5′ GMP production to create thicker and more durable biofilms,74 whereas Pel enhances bacteria tolerance to aminoglycoside antibiotics and antibiotic colistin.9,75 Both the polysaccharides protect bacteria embedded in biofilm from neutrophil phagocytosis and antimicrobials, creating a powerful defense strategy for the progression of the infection.73,76,77 However, Pseudomonas strain-specific Psl and Pel switch synthesis depend on the environmental conditions of the wound. Alginate, an acetylated linear, unbranched biopolymer of mannuronic acid and glucuronic acid residues, is reportedly synthesized due to mucA22 allele mutation in mucoid strains. Alginate contributes to the maturation of biofilm, protection from phagocytosis and retention of water and nutrients.78,79,80
Extracellular DNA (eDNA) released into the biofilm due to cell lysis is one of the important components of the biofilm matrix with many functions. It is a nutrient source to bacteria in biofilms, acts as an interconnecting compound for the formation of microbial aggregates in biofilm maturation, facilitates twitching motility for biofilm expansion and lastly creates an acidic environment in biofilms, thereby limiting the entry of antimicrobial agents.81-83 The extracellular appendages of P. aeruginosa, such as the flagella, type IV pili and fimbriae, in addition to motility, also function as adhesives in the interactions between cells and surfaces as well as in the development of microcolonies in biofilms.84,85
Quorum sensing mechanism and biofilm formation
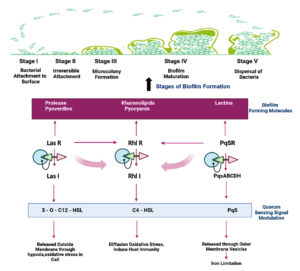
Quorum sensing is another crucial P. aeruginosa regulation mechanism that monitors population size by creating and detecting diffusible signal molecules that control the motility of the organism, synthesis of virulence factors and biofilm formation. Auto signalling synthases activate three main QS systems-Las, Rhl and PQS systems via lactone signaling molecules (3O-C12-HSL, C4-HSL, 2-heptyl-3-hydroxy-4-quinolone). They trigger the synthesis of functional elements such as rhamnolipid, pyocyanin, pyoverdine, Pel polysaccharides and lectins.76,86 Rhamnolipid, a biosurfactant, maintains the pores and channels between microcolonies so that liquid and nutrients can pass through mature biofilms.87 Pyoverdine can bind and transport iron, which is essential for the development of biofilms.88 The cytotoxic compound pyocyanin lyses cells and releases eDNA, increasing the fluid’s viscosity and physicochemical interactions between the biofilm matrix and its environment. It also encourages cellular aggregation.89 Pel polysaccharides interact with eDNA through anionic-cationic interactions to strengthen the biofilm.88 LecA and lecB, two soluble proteins with adhesive capabilities, enable adhesion to biological surfaces and the retention of both cells and exopolysaccharides in a growing biofilm.90,91 In addition, rhl regulation of swarming and twitching motilities in bacterial translocation has been reported to be an important indirect relationship between biofilm development and QS. In the presence of glutamate or succinate as a carbon source, swarming motility results in flat, uniform biofilms.92 Twitching motility, a flagella-independent translocation, resulting in forming P. aeruginosa microcolonies in iron-limited media.93 Together with other polymeric elements, these molecular and biological interactions lead to the formation of a developed and robust biofilm (Figure 2).
Figure 2. Schematic illustration of biofilm formation in Pseudomonas aeruginosa through three main quorum sensing systems-Las, Rhl and PQS systems
Current Pseudomonas specific therapeutic strategies in controlling diabetic foot ulcer
Biofilms are critical in DFU because the colonizing bacteria interact to generate a synergistic environment conducive for infection progression and as a result, the formation of a chronic wound. Due to ineffective antibiotic therapy, alternatively that has gained the interest of researches is biofilm-based wound therapy. The first step in this therapy is the degradation of the biofilms, followed by the application of antimicrobial drugs to kill or inhibit microorganisms embedded in the wound. As the biofilm bioburden level decreases, so does the inflammatory response (neutrophils and macrophages), proteases and reactive oxygen level decreases. As a result, the wound will transition from a chronic to an active healing condition. This results in active healing of the wound. In 2017, World Health Organization identified P. aeruginosa as one of the most harmful bacteria and categorized it as a priority pathogen for the development of new targeted drug deliveries. Current DFU therapies still in clinical trials mainly focus on controlling Pseudomonas biofilm formation, inhibiting quorum sensing pathway essential for biofilm formation and other specific enzymes to develop potent antimicrobial therapeutics. Table 2 summarizes therapeutics against P. aeruginosa in diabetic foot ulcers.
Table (2):
List of therapeutics, their advantages and limitations targeting Pseudomonas aeruginosa in diabetic foot ulcers
| Therapeutics | Activity | Advantages | Limitations | Examples | Reference |
|---|---|---|---|---|---|
| Antimicrobial peptides | •Antibiofilm • Anti-Quorum sensing |
• Interact and penetrate the bacterial cell membrane to cause the death of the bacteria • Broad-spectrum microbicidal activity |
• Toxic • Short half-life • Haemolytic activity • Expensive |
LL-37, P5, cationic peptide 1,037, MC1, WLBU2, Fowlicidin-1, BMAP-27, Protegrin PG-5. | [93, 94] |
| Natural products | Antibiofilm agents | • C-glycosidic inhibitors of lectin. • Interferes with the quorum sensing and signalling pathways. |
• Complex extraction process • Low availability • Low penetration into biofilm • Low bactericidal activity |
Dimethylthiophene 22 (sulphonamide), phenylacetylene bearing thiophene, Ajoene, T6SS, norbgugaine, baicalin.
|
[95,97] |
| Anti-Quorum sensing agents | • Broad-spectrum quorum sensing pathway inhibitors • Inhibit Las A protease and rhamnolipid production. |
||||
| Chemicals | • Gallium disrupts bacterial Fe metabolism and inhibit P. aeruginosa growth • NO creates nitrosative stress or oxidative stress in the biofilm and aids in biofilm dispersal |
• Cytotoxicity • Resistant development |
Gallium, dimethylthiophene 22 (sulphonamide), phenylacetylene bearing thiophene | [93] | |
| Bacteriophages | • Narrow spectrum of activity • Safer and better tolerated as they can replicate within target bacterium without infecting mammalian cells • Easy administration • Less expensive • Could be engineered to achieve better efficacy. |
• Absence of specificity towards a bacterial strain • Formulation and stabilization of pharmaceutical product is difficult • Gradual emergence of bacterial resistance against bacteriophages. • Bacteriophages may also contribute to development of antibiotic resistance |
IME180 | [98] | |
| Nanoparticles | • Produce reactive oxygen species (ROS) that disrupt replication of the organism.
• Inhibit biofilm formation pathways. |
• There is no standard testing method for evaluating the antimicrobial activity of the synthesized nanoparticles. • There is no homogeneous culture medium used among different research communities working with the synthesis of nanoparticle base anti microbials. |
Piper betel (Pb) mediated AgNPs (Pb-AgNPs), Zinc Oxide Nano particles, copper nanoparticles (CuNPs). | [99-102] |
Potential therapeutics used for dealing antibiotic resistance in Pseudomonas aeruginosa
Anti-biofilm therapeutics
Enzymatic and synthetic therapeutics are being employed to degrade and disperse biofilms for controlling Pseudomonas infections. Microbial enzymes such as alginate lyase,103 and glucosyl hydrolases (dextranase and mutanase),104,105 were therapeutically being used singly or in combination with other antibiotics targeting exopolysaccharide polymers and alginate components of the biofilm. Deoxyribonuclease (DNAase) extracted from the human eye were applied for the degradation eDNA.106 Naturally, Ginger (Zingiber officinale Rosc) extracts were reported to inhibit biofilm formation by decreasing the production of cyclic 3’5′ GMP.107 Similarly, ethanol extracts containing casbane diterpene from Croton nepetaefolius Baill plant were found to inhibit biofilm formation by interacting with lipopolysaccharides of cell membranes.108 Marine sponges (Agelas conifer, Agelaceae) synthesize pyrrole-imidazole alkaloids bromoageliferin, which has been shown to prevent the growth of new biofilms and disperse existing ones.109 Sulphonamides such as dimethylthiophene have potent anti-biofilm activity and are C-glycoside inhibitors with high affinity towards lecB protein of Pseudomonas.96 Additionally, low dose of nitric oxide gas was found to disperse biofilm and expose microbial species to antibiotics.110 Temporin B from frogs,111 indolicidin from the cytoplasmic granules of bovine neutrophils.112 Human beta-defensin 3,113 and many others have been reported to have anti-biofilm activities. Anti-microbial peptides could be promising alternative therapeutics to deal with antimicrobial resistance in the future.
Anti- Quorum sensing therapeutics
Quorum sensing is a desirable target for biofilm suppression and removal. It was discovered that the naturally available carotenoid zeaxanthin targets the Las and Rhl system and inhibits biofilm.114 The plant flavonoid quercetin is well known for its pharmacological activities, which include lowering pyocyanin synthesis and preventing P. aeruginosa from forming biofilms.115 P. aeruginosa periplasmic enzyme PvdQ acylase, another quorum-suppressing compound, has been shown to hydrolyze N-acyl homoserine lactone (AHL), reducing virulence and easing infections.116 The fungal metabolite, terrein, derived from Aspergillus terreus, has been shown to inhibit both QS system and cyclic 3’5′ GMP without impacting bacterial survival.117 Quenching enzymes, QsdA and AqdC isolated from Rhodococcus erythropolis decreased N-acylhomoserine lactone synthesis, inhibiting bioactive compounds required for biofilm formation. Ajoene, a sulfur rich QS targetting molecule derived from garlic that targets RsmY and RsmZ in P. aeruginosa.95 Similarly, baicalin flavonoid purified from the roots of Scutellaria baicalensis, repressed QS-regulatory genes, including lasI, lasR, rhlI, rhlR, pqsR and pqsA in P. aeruginosa and minimised the virulence phenotypes such as LasA protease, LasB elastase, pyocyanin, rhamnolipid, motilities and exotoxin A.97
Therapeutics against iron metabolism
P. aeruginosa acquires extracellular iron via iron absorption mechanisms (siderophores). Therefore, iron analogues and chelators that target iron metabolism may be effective treatments for P. aeruginosa infections. Gallium, which resembles iron structurally, was used as an alternative to iron to obstruct iron uptake, obstruct iron-dependent pathways, affect bacterial survival, and obstruct biofilm formation.118 Deferoxamine and deferasirox, two FDA-approved iron chelation compounds, are used with tobramycin to effectively break up existing biofilms.119
Fifth generation antibiotics
Due to high resistance to conventional antibiotics, fifth-generation antibiotics are used singly or in combination, specifically against Pseudomonas species. Cephalosporin or tazobactam was effective against Gram-negative bacteria and being used as an antipseudomonal agent.120 A combination of ceftolozane-tazobactam was reported to be effective in the downregulation of ampC gene, against the adhesion of colonies to the surface and biofilm formation.121,122 Similarly, ureidopenicillin, a beta-lactamase inhibitor found to be effective against P. aeruginosa.121,123
Immunotherapeutics
Monoclonal antibodies (mABs) targeting bacterial DNA binding proteins have been emerging as a promising therapeutic tool against P. aeruginosa in mouse models.124 KaloBios designed KB001-A, an anti-P. aeruginosa mAb against response to the Type III secretion system (T3SS), which is necessary for P. aeruginosa pathogenicity and was found to be safe and well-tolerated.8,125 Many monoclonal antibodies including MEDI3902 from AstraZeneca targeting Pseudomonas biofilms are in clinical trials.126
Nanoparticles
Nanoparticles have been employed for the penetration of antibiotics into biofilms. For instance, it has been demonstrated that clinical P. aeruginosa strains resistant to specific antibiotics are susceptible to the antibacterial action of silver nanoparticles.127 Without altering the development of planktonic cells, zinc ions and ZnO nanoparticles have been shown to prevent the formation of biofilms and also inhibit pyocyanin, pyochelin and hemolytic activity.100 Methanolic silver nanoparticles showed 85.63% inhibition of Pseudomonas biofilm formation.99 TTO (Tea of Tree oil) nanoparticles has potential antibiofilm activity against P. aeruginosa PAO1,128 and D-galactose nanoparticles could inhibit lecA of Pseudomonas significantly.129 Additionally, polyphosphoester nanoparticles, silver acetate, and minocycline significantly improved P. aeruginosa’s susceptibility.130
Diabetes foot ulcers can result in lower limb amputations and significantly negatively impact the socioeconomic and health of diabetic patients. Chronic DFU wounds are polymicrobial with varied organisms. Pseudomonas aeruginosa is the dominant pathogen present in medium and chronic diabetic foot ulcers. It is regarded as a highly harmful pathogen due to its ability to form multidrug resistance biofilms. The extraordinary capacity of P. aeruginosa to create biofilms is aided by a highly developed quorum-sensing cell communication system and the activation of antibiotic resistance pathways. Nowadays, several therapeutic strategies are developing to prevent resistance such as combination therapy by combining antibiotics of different classes, discovery of novel antibiotics such as ceftolozane-tazobactam, ceftazidime-avibactam, along with designing of novel anti-microbial peptides could be highly promising against multidrug resistant Pseudomonas aeruginosa. Future research is needed to create more sophisticated methods that can provide high-throughput and precise treatment at an early stage of P. aeruginosa proliferation and biofilm formation.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DC conceptualized the idea, performed literature review, and wrote the manuscript. SK critically reviewed and revised the manuscript. Both authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Mariadoss AVA, Sivakumar AS, Lee C-H, Kim SJ. Diabetes mellitus and diabetic foot ulcer: Etiology, biochemical and molecular based treatment strategies via gene and nanotherapy. Biomed Pharmacother. 2022;151:113-134.
Crossref - Sun H, Saeedi P, Karuranga S, et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183:109119.
Crossref - Pradeepa R, Mohan V. Epidemiology of type 2 diabetes in India. Indian J Ophthalmol. 2021;69(11):2932-2938.
Crossref - Gregg EW, Sattar N, Ali MK. The changing face of diabetes complications. Lancet Diabetes Endocrinol. 2016;4(6):537-547.
Crossref - Tomic D, Shaw JE, Magliano DJ. The burden and risks of emerging complications of diabetes mellitus. Nat Rev Endocrinol. 2022;18(9):525-539.
Crossref - Shahbazian H, Yazdanpanah L, Latifi SM. Risk assessment of patients with diabetes for foot ulcers according to risk classification consensus of international working group on diabetic foot (IWGDF). Pak J Med Sci. 2013;29(3):730-734.
Crossref - Raja JM, Maturana MA, Kayali S, Khouzam A, Efeovbokhan N. Diabetic foot ulcer: A comprehensive review of pathophysiology and management modalities. World J Clin Cases. 2023;11(8):1684-1693.
Crossref - Yang L, Rong G-C, Wu Q-N. Diabetic foot ulcer: Challenges and future. World J Diabetes. 2022;13(12):1014-1034.
Crossref - Prompers L, Huijberts M, Apelqvist J, et al. High prevalence of ischaemia, infection and serious comorbidity in patients with diabetic foot disease in Europe. Baseline results from the Eurodiale study. Diabetologia. 2007;50(1):18-25.
Crossref - Leone S, Pascale R, Vitale M, Esposito S. [Epidemiology of diabetic foot]. Infez Med. 2012;20(Suppl 1):8-13.
- Girijala RL, Bush RL. Review of Socioeconomic Disparities in Lower Extremity Amputations: A Continuing Healthcare Problem in the United States. Cureus. 2018;10(10):e3418.
Crossref - Skrepnek GH, Armstrong DG, Mills JL. Open bypass and endovascular procedures among diabetic foot ulcer cases in the United States from 2001 to 2010. Journal of Vascular Surgery. 2014;60(5):1255-1265.
Crossref - Cavanagh P, Attinger C, Abbas Z, Bal A, Rojas N, Xu ZR. Cost of treating diabetic foot ulcers in five different countries: Cost of Treating Diabetic Foot Ulcers in Five Countries. Diabetes Metab Res Rev. 2012;28(Suppl 1):107-111.
Crossref - Ghosh P, Valia R. Burden of Diabetic Foot Ulcers in India: Evidence Landscape from Published Literature. Value in Health. 2017;20(9):A485.
Crossref - Moore Z, Avsar P, Wilson P, et al. Diabetic foot ulcers: treatment overview and cost considerations. J Wound Care. 2021;30(10):786-791.
Crossref - Khanolkar MP, Bain SC, Stephens JW. The diabetic foot. QJM. 2008;101(9):685-695.
Crossref - Bandyk DF. The diabetic foot: Pathophysiology, evaluation, and treatment. Semin Vasc Surg. 2018;31(2-4):43-48.
Crossref - Deng H, Li B, Shen Q, et al. Mechanisms of diabetic foot ulceration: A review. J Diabetes. 2023;15(4):299-312.
Crossref - Noor S, Zubair M, Ahmad J. Diabetic foot ulcer-A review on pathophysiology, classification and microbial etiology. Diabetes Metab Syndr. 2015;9(3):192-199.
Crossref - Snyder RJ, Hanft JR. Diabetic foot ulcers–effects on QOL, costs, and mortality and the role of standard wound care and advanced-care therapies. Ostomy Wound Manage. 2009;55(11):28-38
- Basra R, Papanas N, Farrow F, Karalliedde J, Vas P. Diabetic Foot Ulcers and Cardiac Autonomic Neuropathy. Clin Ther. 2022;44(2):323-330.
Crossref - Chatwin KE, Abbott CA, Boulton AJM, Bowling FL , Reeves ND. The role of foot pressure measurement in the prediction and prevention of diabetic foot ulceration-A comprehensive review. Diabetes Metab Res Rev. 2020;36(4):e3258.
Crossref - Richard C, Wadowski M, Goruk S, Cameron L, Sharma AM, Field CJ. Individuals with obesity and type 2 diabetes have additional immune dysfunction compared with obese individuals who are metabolically healthy. BMJ Open Diabetes Res Care. 2017;5(1):e000379.
Crossref - Rosyid FN. Etiology, pathophysiology, diagnosis and management of diabetics’ foot ulcer. Int J Res Med Sci. 2017;5(10):4206-4213.
Crossref - Dorr S, Holland-Letz A-K, Weisser G, Chatzitomaris A, Lobmann R. Bacterial Diversity, Antibiotic Resistance, and the Risk of Lower Limb Amputation in Younger and Older Individuals With Diabetic Foot Infection. Int J Low Extrem Wounds. 2023;22(1):63-71.
Crossref - Chai W, Wang Y, Zheng H, et al. The Profile of Microbiological Pathogens in Diabetic Foot Ulcers. Front Med. 2021;8:656467.
Crossref - Sadeghpour Heravi F, Zakrzewski M, Vickery K, Armstrong DG, Hu H. Bacterial Diversity of Diabetic Foot Ulcers: Current Status and Future Prospectives. J Clin Med. 2019;8(11):1935.
Crossref - Wilmes P, Bond PL. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14(2):92-97.
Crossref - Lipsky BA, Berendt AR, Cornia PB, et al. 2012 Infectious Diseases Society of America Clinical Practice Guideline for the Diagnosis and Treatment of Diabetic Foot Infectionsa. Clin Infect Dis. 2012;54(12):e132-e173.
Crossref - Banu A, Noorul Hassan MM, Rajkumar J, Srinivasa S. Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: A prospective study. Australas Med J. 2015;8(9):280-285.
Crossref - Tascini C, Piaggesi A, Tagliaferri E, et al. Microbiology at first visit of moderate-to-severe diabetic foot infection with antimicrobial activity and a survey of quinolone monotherapy. Diabetes Res Clin Pract. 2011;94(1):133-139.
Crossref - Waldman O, Sajda T, Oh I, Sulovari A. Group B Streptococcus Infected Tenosynovitis in Diabetic Foot Ulcers. Foot Ankle Orthop. 2020;5(4):2473011420S0048.
Crossref - Jneid J, Cassir N, Schuldiner S, et al. Exploring the Microbiota of Diabetic Foot Infections With Culturomics. Front Cell Infect Microbiol. 2018;8:282.
Crossref - Benwan KA, Mulla AA, Rotimi VO. A study of the microbiology of diabetic foot infections in a teaching hospital in Kuwait. J Infect Public Health. 2012;5(1):1-8.
Crossref - Sivanmaliappan TS, Sevanan M. Antimicrobial Susceptibility Patterns of Pseudomonas aeruginosa from Diabetes Patients with Foot Ulcers. Int J Microbiol. 2011;605195.
Crossref - Zakhour J, Sharara SL, Hindy J-R, Haddad SF, Kanj SS. Antimicrobial Treatment of Pseudomonas aeruginosa Severe Sepsis. Antibiotics. 2022;11(10):1432.
Crossref - Charles PGP, Uckay I, Kressmann B, Emonet S, Lipsky BA. The role of anaerobes in diabetic foot infections. Anaerobe. 2015;34:8-13.
Crossref - Zubair M, Malik A, Ahmad J. Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in North India. The Foot. 2011;21(1):6-14.
Crossref - Shankar EM, Mohan V, Premalatha G, Srinivasan RS, Usha AR. Bacterial etiology of diabetic foot infections in South India. Eur J Intern Med. 2005;16(8):567-570.
Crossref - Jain S, Barman R. Bacteriological profile of diabetic foot ulcer with special reference to drug-resistant strains in a tertiary care center in North-East India. Indian J Endocr Metab. 2017;21(5):688-694.
Crossref - Otta S, Debata N, Swain B. Bacteriological profile of diabetic foot ulcers. CHRISMED J Health Res. 2019;6(1):7-11.
Crossref - Mehta V, Kikani K, Mehta S. Microbiological profile of diabetic foot ulcers and its antibiotic susceptibility pattern in a teaching hospital, Gujarat. Int J Basic Clin Pharmacol. 2014;3(1):92.
Crossref - Kaimkhani GM, Siddiqui AA, Rasheed N, et al. Pattern of Infecting Microorganisms and Their Susceptibility to Antimicrobial Drugs in Patients with Diabetic Foot Infections in a Tertiary Care Hospital in Karachi, Pakistan. Cureus. 2018;10(6):e2872.
Crossref - Jneid J, Lavigne JP, La Scola B, Cassir N. The diabetic foot microbiota: A review. Human Microbiome Journal. 2017;5-6:1-6.
Crossref - Suryaletha K, John J, Radhakrishnan MP, George S, Thomas S. Metataxonomic approach to decipher the polymicrobial burden in diabetic foot ulcer and its biofilm mode of infection. Int Wound J. 2018;15(3):473-481.
Crossref - Citron DM, Goldstein EJC, Merriam CV, Lipsky BA, Abramson MA. Bacteriology of Moderate-to-Severe Diabetic Foot Infections and In Vitro Activity of Antimicrobial Agents. J Clin Microbiol. 2007;45(9):2819-2828.
Crossref - Watters C, DeLeon K, Trivedi U, et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol. 2013;202(2):131-141.
Crossref - Afonso AC, Oliveira D, Saavedra MJ, Borges A, Simoes M. Biofilms in Diabetic Foot Ulcers: Impact, Risk Factors and Control Strategies. Int J Mol Sci. 2021;22(15):8278.
Crossref - Preda VG, Sandulescu O. Communication is the key: biofilms, quorum sensing, formation and prevention. Discoveries (Craiova). 2019;7(3):e100.
Crossref - Mashaly M, Abo El kheir M, Ibrahim M, Khafagy W. Aerobic bacteria isolated from diabetic foot ulcers of Egyptian patients: types, antibiotic susceptibility pattern and risk factors associated with multidrug-resistant organisms. Germs. 2021;11(4):570-582.
Crossref - Oh KY, Lee S, Lee M-S, et al. Composition of Vaginal Microbiota in Pregnant Women With Aerobic Vaginitis. Front Cell Infect Microbiol. 2021;11:677648.
Crossref - Glen KA, Lamont IL. β-lactam Resistance in Pseudomonas aeruginosa: Current Status, Future Prospects. Pathogens. 2021;10(12):1638.
Crossref - Balasubramanian D, Kumari H, Mathee K. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathogens Disease. 2014;73(2):1-14.
Crossref - Bush K, Jacoby GA. Updated Functional Classification of β-Lactamases. Antimicrob Agents Chemother. 2010;54(3):969-976.
Crossref - Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β -lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrobial Resistance. 2021;3(3):dlab092.
Crossref - Lin H, Feng C, Zhu T, et al. Molecular Mechanism of the β-Lactamase Mediated β-Lactam Antibiotic Resistance of Pseudomonas aeruginosa Isolated From a Chinese Teaching Hospital. Front Microbiol. 2022;13:855961.
Crossref - Ahmadian L, Norouzi Bazgir Z, Ahanjan M, Valadan R, Goli HR. Role of Aminoglycoside-Modifying Enzymes (AMEs) in Resistance to Aminoglycosides among Clinical Isolates of Pseudomonas aeruginosa in the North of Iran. BioMed Res Int. 2021;1-10.
Crossref - Thacharodi A, Lamont IL. Aminoglycoside-Modifying Enzymes Are Sufficient to Make Pseudomonas aeruginosa Clinically Resistant to Key Antibiotics. Antibiotics. 2022;11(7):884.
Crossref - Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. IJMS. 2022;23(24):15779.
Crossref - Pang Z, Raudonis R, Glick BR, et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Kunz Coyne AJ, El Ghali A, Holger D, Rebold N, Rybak MJ. Therapeutic Strategies for Emerging Multidrug-Resistant Pseudomonas aeruginosa. Infect Dis Ther. 2022;11(2):661-682.
Crossref - Dreier J, Ruggerone P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front Microbiol. 2015;6:660.
Crossref - Goli HR, Nahaei MR, Rezaee MA, et al. Role of MexAB-OprM and MexXY-OprM efflux pumps and class 1 integrons in resistance to antibiotics in burn and Intensive Care Unit isolates of Pseudomonas aeruginosa. J Infect Public Health. 2018;11(3):364-372.
Crossref - Llanes C, Kohler T, Patry I, Dehecq B, van Delden C, Plesiat P. Role of the MexEF-OprN Efflux System in Low-Level Resistance of Pseudomonas aeruginosa to Ciprofloxacin. Antimicrob Agents Chemother. 2011;55(12):5676-5684.
Crossref - Alcalde-Rico M, Olivares-Pacheco J, Alvarez-Ortega C, Camara M, Martinez JL. Role of the Multidrug Resistance Efflux Pump MexCD-OprJ in the Pseudomonas aeruginosa Quorum Sensing Response. Front Microbiol. 2018;9:2752.
Crossref - Shigemura K, Osawa K, Kato A, et al. Association of overexpression of efflux pump genes with antibiotic resistance in Pseudomonas aeruginosa strains clinically isolated from urinary tract infection patients. J Antibiot. 2015;68(9):568-572.
Crossref - Sada M, Kimura H, Nagasawa N, et al. Molecular Evolution of the Pseudomonas aeruginosa DNA Gyrase gyrA Gene. Microorganisms. 2022;10(8):1660.
Crossref - Tian Z-X, Wang Y-P. Identification of cpxS mutational resistome in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2023;67(11).
Crossref - Poole K. Pseudomonas Aeruginosa: Resistance to the Max. Front Microbiol. 2011;2:65.
Crossref - Ghafoor A, Hay ID, Rehm BHA. Role of Exopolysaccharides in Pseudomonas aeruginosa Biofilm Formation and Architecture. Appl Environ Microbiol. 2011;77(15):5238-5246.
Crossref - Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10(6):644-648.
Crossref - Byrd MS, Sadovskaya I, Vinogradov E, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73(4):622-638.
Crossref - Colvin KM, Alnabelseya N, Baker P, Whitney JC, PHowel lL, Matthew R Parsek. PelA Deacetylase Activity Is Required for Pel Polysaccharide Synthesis in Pseudomonas aeruginosa. J Bacteriol. 2013;195(10):2329-2339.
Crossref - Irie Y, Borlee BR, O’Connor JR, et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109(50):20632-20636.
Crossref - Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development: EPS-mediated biofilm development. Environ Microbiol. 2011;13(7):1705-1717.
Crossref - Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa Biofilms. Int J Mol Sci. 2020;21(22):8671.
Crossref - Billings N, Ramirez Millan M, Caldara M, et al. The Extracellular Matrix Component Psl Provides Fast-Acting Antibiotic Defense in Pseudomonas aeruginosa Biofilms. PLoS Pathog. 2013;9(8):e1003526.
Crossref - Hay ID, Rehman ZU, Moradali MF, Wang Y, Rehm BHA. Microbial alginate production, modification and its applications. Microb Biotechnol. 2013;6(6):637-650.
Crossref - Singh S, Datta S, Narayanan KB, Rajnish KN. Bacterial exo-polysaccharides in biofilms: role in antimicrobial resistance and treatments. J Genet Eng Biotechnol. 2021;19(1):140.
Crossref - Wloka M, Rehage H, Flemming H-C, Wingender J. Structure and rheological behaviour of the extracellular polymeric substance network of mucoid Pseudomonas aeruginosa biofilms. Biofilms. 2005;2(4):275-283.
Crossref - Wilton M, Charron-Mazenod L, Moore R, Lewenza S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(1):544-553.
Crossref - Gloag ES, Turnbull L, Huang A, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA. 2013;110(28):11541-11546.
Crossref - Mulcahy H, Charron-Mazenod L, Lewenza S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol. 2010;12(6):1621-1629.
Crossref - Barken KB, Pamp SJ, Yang L, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10(9):2331-2343.
Crossref - Beaussart A, Baker AE, Kuchma SL, El-Kirat-Chatel S, O’Toole GA, Dufrene YF. Nanoscale Adhesion Forces of Pseudomonas aeruginosa Type IV Pili. ACS Nano. 2014;8(10):10723-10733.
Crossref - Nadal Jimenez P, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76(1):46-65.
Crossref - Pamp SJ, Tolker-Nielsen T. Multiple Roles of Biosurfactants in Structural Biofilm Development by Pseudomonas aeruginosa. J Bacteriol. 2007;189(6):2531-2539.
Crossref - Sakuragi Y, Kolter R. Quorum-Sensing Regulation of the Biofilm Matrix Genes ( pel ) of Pseudomonas aeruginosa. J Bacteriol. 2007;189(14):5383-5386.
Crossref - Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin Facilitates Extracellular DNA Binding to Pseudomonas aeruginosa Influencing Cell Surface Properties and Aggregation. PLoS ONE. 2013;8(3):e58299.
Crossref - Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8(6):1095-1104.
Crossref - Passos da Silva D, Matwichuk ML, Townsend DO, et al. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat Commun. 2019;10(10):2183.
Crossref - Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62(5):1264-1277.
Crossref - Yin R, Cheng J, Wang J, Li P, Lin J. Treatment of Pseudomonas aeruginosa infectious biofilms: Challenges and strategies. Front Microbiol. 2022;13:955286.
Crossref - Lei J, Sun L, Huang S, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11(7):3919-3931
- Jakobsen TH, Warming AN, Vejborg RM, et al. A broad range quorum sensing inhibitor working through sRNA inhibition. Sci Rep. 2017;7(1):9857.
Crossref - Sommer R, Rox K, Wagner S, et al. Anti-biofilm Agents against Pseudomonas aeruginosa : A Structure-Activity Relationship Study of C -Glycosidic LecB Inhibitors. J Med Chem. 2019;62(20):9201-9216.
Crossref - Luo J, Dong B, Wang K, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE. 2017;12(4):e0176883.
Crossref - Principi N, Silvestri E, Esposito S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front Pharmacol. 2019;10:513.
Crossref - LewisOscar F, Nithya C, Vismaya S, et al. In vitro analysis of green fabricated silver nanoparticles (AgNPs) against Pseudomonas aeruginosa PA14 biofilm formation, their application on urinary catheter. Progress in Organic Coatings. 2021;151:106058.
Crossref - Lee J-H, Kim Y-G, Cho MH, Lee J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiological Research. 2014;169(12):888-896.
Crossref - Shah S, Gaikwad S, Nagar S, et al. Biofilm inhibition and anti-quorum sensing activity of phytosynthesized silver nanoparticles against the nosocomial pathogen Pseudomonas aeruginosa. Biofouling. 2019;35(1):34-49.
Crossref - Duval RE, Gouyau J, Lamouroux E. Limitations of Recent Studies Dealing with the Antibacterial Properties of Silver Nanoparticles: Fact and Opinion. Nanomaterials. 2019;9(12):1775.
Crossref - Lamppa JW, Griswold KE. Alginate Lyase Exhibits Catalysis-Independent Biofilm Dispersion and Antibiotic Synergy. Antimicrob Agents Chemother. 2013;57(1):137-145.
Crossref - Pleszczynska M, Wiater A, Janczarek M, Szczodrak J. (1→3)-α-d-Glucan hydrolases in dental biofilm prevention and control: A review. International Journal of Biological Macromolecules. 2015;79:761-778.
Crossref - Boddapati S, Gummadi SN. Production and application of purified mutanase from novel Cellulosimicrobium funkei SNG1 in invitro biofilm degradation. Biotech App Biochem. 2023;70(3):1371-1383.
Crossref - Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and Antibiotics on Biofilm Characteristics. Antimicrob Agents Chemother. 2009;53(3):1204-1209.
Crossref - Kim H-S, Park H-D. Ginger Extract Inhibits Biofilm Formation by Pseudomonas aeruginosa PA14. PLoS ONE. 2013;8(9):e76106.
Crossref - Carneiro VA, Santos HS dos, Arruda FVS, et al. Casbane Diterpene as a Promising Natural Antimicrobial Agent against Biofilm-Associated Infections. Molecules. 2010;16(1):190-201.
Crossref - Huigens RW, Richards JJ, Parise G, et al. Inhibition of Pseudomonas aeruginosa Biofilm Formation with Bromoageliferin Analogues. J Am Chem Soc. 2007;129(22):6966-6967.
Crossref - Howlin RP, Cathie K, Hall-Stoodley L, et al. Low-Dose Nitric Oxide as Targeted Anti-biofilm Adjunctive Therapy to Treat Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. Molecular Therapy. 2017;25(9):2104-2116.
Crossref - Simmaco M, Mignogna G, Canofeni S, Miele R, Mangoni ML, Barra D. Temporins, Antimicrobial Peptides from the European Red Frog Rana temporaria. Eur J Biochem. 1996;242(3):788-792.
Crossref - de la Fuente-Nunez C, Hancock REW. Using anti-biofilm peptides to treat antibiotic-resistant bacterial infections. PDJ. 2015;3(2):1-8.
Crossref - Huang LC, Redfern RL, Narayanan S, Reins RY, McDermott AM. In vitro activity of human beta-defensin 2 against Pseudomonas aeruginosa in the presence of tear fluid. Antimicrob Agents Chemother. 2007;51(11):3853-3860.
Crossref - Lin Chua S, Liu Y, Li Y, et al. Reduced Intracellular c-di-GMP Content Increases Expression of Quorum Sensing-Regulated Genes in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2017;7:451.
Crossref - Ouyang J, Sun F, Feng W, et al. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol. 2016;120(4):966-974.
Crossref - Utari PD, Setroikromo R, Melgert BN, Quax WJ. PvdQ Quorum Quenching Acylase Attenuates Pseudomonas aeruginosa Virulence in a Mouse Model of Pulmonary Infection. Front Cell Infect Microbiol. 2018;8:119.
Crossref - Kim B, Park J-S, Choi H-Y, Yoon SS, Kim W-G. Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP. Sci Rep. 2018;8:8617.
Crossref - Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest. 2007;117(4):877-888.
Crossref - Moreau-Marquis S, O’Toole GA, Stanton BA. Tobramycin and FDA-Approved Iron Chelators Eliminate Pseudomonas aeruginosa Biofilms on Cystic Fibrosis Cells. Am J Respir Cell Mol Biol. 2009;41(3):305-313.
Crossref - Srivastava P, Sivashanmugam K. Efficacy of sub-MIC level of meropenem and ciprofloxacin against extensive drug-resistant (XDR) Pseudomonas aeruginosa isolates of diabetic foot ulcer patients. Infect Genet Evol. 2021;92:104824.
Crossref - Li C, Nicolav DP, Lister PD, Quimtiliani R, Nightingale CH. Pharmacodynamic study of -lactams alone and in combination with -lactamase inhibitors against Pseudomonas aeruginosa possessing an inducible -lactamase. J Antimicrob Chemother. 2004;53(2):297-304.
Crossref - Cluck D, Lewis P, Stayer B, Spivey J, Moorman J. Ceftolozane-tazobactam: A new-generation cephalosporin. Am J Health-Syst Pharm. 2015;72(24):2135-2146.
Crossref - Zelenitsky S, Nash J, Weber Z, Iacovides H, Ariano R. Targeted benefits of prolonged-infusion piperacillin-tazobactam in an in vitro infection model of Pseudomonas aeruginosa. J Chemother. 2016;28(5):390-394.
Crossref - Novotny LA, Jurcisek JA, Goodman SD, Bakaletz LO. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. eBioMedicine. 2016;10:33-44.
Crossref - Jain R, Beckett V, Konstan M, et al. KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J Cyst Fibros. 2017;17(4):484-491:.
Crossref - Ali SO, Yu XQ, Robbie GJ, et al. Phase 1 study of MEDI3902, an investigational anti-Pseudomonas aeruginosa PcrV and Psl bispecific human monoclonal antibody, in healthy adults. Clin Microbiol Infect. 2019;25(5):629.e1-629.e6.
Crossref - Salomoni R, Leo P, Montemor A, Rinaldi BG , Rodrigues M. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol Sci Appl. 2017;10:115-121.
Crossref - Comin VM, Lopes LQS, Quatrin PM, et al. Influence of Melaleuca alternifolia oil nanoparticles on aspects of Pseudomonas aeruginosa biofilm. Microbial Pathogenesis. 2016;93:120-125.
Crossref - Flockton T, Schnorbus L, Araujo A, Adams J, Hammel M, Perez LJ . Inhibition of Pseudomonas aeruginosa Biofilm Formation with Surface Modified Polymeric Nanoparticles. Pathogens. 2019;8(2):55.
Crossref - Chen Q, Shah KN, Zhang F, et al. Minocycline and Silver Dual-Loaded Polyphosphoester-Based Nanoparticles for Treatment of Resistant Pseudomonas aeruginosa. Mol Pharmaceutics. 2019;16(4):1606-1619.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.