ISSN: 0973-7510
E-ISSN: 2581-690X
Conventional blood culture media often fail to isolate Streptococcus pneumoniae, a key cause of pneumonia and bacteremia. This study aimed to improve the detection rate of S. pneumoniae using selective media. The study involved 45 blood samples from patients suspected of bacteremia and pneumonia at the Universitas Sumatera Utara Hospital. The samples were cultured using tryptic soy to make sheep blood 5% with and without gentamicin (TSSBA-G and TSSBA) in detecting S. pneumoniae in the blood of patients with bacteremia and pneumonia. Blood samples were incubated in the BAcT/Alert tube to detect the presence or absence of microorganism growth in blood samples. S. pneumoniae was isolated in 6 of 45 samples (13%) using TSSBA-G. TSSBA without gentamicin yielded 11 cultures (24%) including potential contaminants. Routine media (blood agar and MacConkey agar) detected 12 positive cultures (27%) but failed to isolate S. pneumoniae. Combined use of TSSBA-G and TSSBA increased total positivity to 29/45 (64%). The use of TSSBA-G, especially alongside TSSBA, substantially increases blood culture positivity, particularly for isolating S. pneumoniae a key cause of bacteremia. These findings support the adoption of selective media to improve diagnostic yield.
Streptococcus pneumoniae, TSSBA-G, Blood Culture, Bacteremia, Pneumonia
Streptococcus pneumoniae is a major cause of invasive pneumococcal disease (IPD), which includes bacteremia, meningitis, and pneumonia. Pneumococcal bacteremia, in particular, contributes significantly to global morbidity and mortality. A systematic review and meta-analysis reported an overall mortality rate of 20.8% for IPD, with higher rates observed in elderly populations. IPD is defined by the presence of S. pneumoniae in normally sterile sites such as blood, cerebrospinal fluid, or pleural fluid.1,2
Despite its clinical importance, the isolation of S. pneumoniae from blood cultures remains a challenge. Blood culture remains the gold standard for detection of bacteremia caused by S. pneumoniae, but the sensitivity of conventional methods is often suboptimal.3,4 Optimal growth of this facultative anaerobe often depends on using the right media and atmospheric environments (aerobic, anaerobic, or microaerophilic) conditions, which are critical to enhancing the recovery and identification of the organism.5-7
The bacterium’s comparatively slow growth rate under normal circumstances and the interference of another microorganism with a faster growth rate are two of the many reasons that contribute to this poor detection rate. Another important factor is that prior antibiotic treatment can significantly reduce the likelihood of successfully isolating S. pneumoniae from blood samples.8,9 Previous reports indicate that pneumococcal isolation rates from routine culture systems may fall below 10% in suspected pneumonia and sepsis cases, underscoring the need for more selective culture strategies.10,11
The low sensitivity of blood culture, autolysis, and contamination with fast-growing microorganisms jeopardizes isolation of S. pneumoniae.12,13 Current routine culture media used in Microbiology Laboratories are sheep blood Agar (SBA) and MacConkey agar (MA) for isolation of S. pneumoniae, which has limitations such as a low positivity rate (<10%), causing frequent underdetection of pathogen.14-16 The routine culture media used in microbiology laboratories for the isolation of S. pneumoniae are sheep blood agar (SBA) and MacConkey agar (MA), both of which have limitations, including a low positivity rate of 5%-8%, leading to frequent underdetection of the pathogen.
One promising approach is the addition of gentamicin to tryptic soy agar with 5% sheep blood, known as TSSBA-G. Gentamicin selectively inhibits Gram-negative bacteria without impairing S. pneumoniae growth, thus increasing recovery rates in mixed or contaminated specimens.10,17,18 Recent studies highlight the effectiveness of selective blood agar in improving culture sensitivity and reducing diagnostic delays, especially in high-burden clinical settings.16,19 To address these limitations, there is a pressing need for more selective and sensitive culture media. This study proposes the use of tryptic soy agar supplemented with 5% sheep blood and gentamicin (TSSBA-G) to enhance the recovery of S. pneumoniae from blood samples of patients with suspected bacteremia or pneumonia.
Study setting
This descriptive study compared the culture positivity rates of Streptococcus pneumoniae using routine media and a selective medium—tryptic soy agar with 5% sheep blood, with and without gentamicin (TSSBA and TSSBA-G). Blood samples were collected from hospitalized patients with suspected bacteremia or pneumonia at the Clinical Microbiology Laboratory, Universitas Sumatera Utara Hospital, between January and August 2019. A volume of 5 mL of blood was collected from pediatric patients, and 10 mL from adults. Each blood sample represented an independent clinical case (n = 45); no technical triplicates were performed.
The study protocol was reviewed and approved by the Health Research Ethics Committee of the Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia (Approval No. 11/KEP/USU/2021). Written informed consent was obtained from all participants or their legal guardians prior to sample collection.
Preparation of culture media
The culture media employed in this study included Columbia agar (Oxoid CM0331B), MacConkey agar (Oxoid CM0115B), and Tryptic Soy Agar (TSA; Oxoid CM0131B). Sheep blood, obtained from CV Karya Graha Asri, Jakarta (a certified supplier of laboratory-grade sheep blood), was used for culture preparation.
Routine Media
Blood Agar (BA) was prepared by dissolving 3.8 g of blood agar base in 100 mL of distilled water, followed by autoclaving at 121 °C for 15 minutes. Once cooled to 40-45 °C, 5% defibrinated sheep blood was added, mixed thoroughly, and poured into sterile petri dishes. MacConkey agar was prepared similarly by dissolving 5.15 g of MacConkey agar in 100 mL of distilled water, autoclaving, cooling to 50 °C, and pouring into petri dishes.
Tryptic Soy Sheep Blood Agar (TSSBA) and Tryptic Soy Sheep Blood Agar with Gentamicin (TSSBA-G)
Tryptic soy agar (4 g) was dissolved in 100 mL of distilled water, sterilized by autoclaving, and cooled to 40-45 °C. A 2.5 mg/mL gentamicin stock solution was prepared by dissolving 80 mg of gentamicin in 32 mL of sterile distilled water, filtered, and stored at -20 °C to -70 °C. To prepare TSSBA-G, 1 mL of gentamicin stock was added to 500 mL of cooled tryptic soy agar, along with 5% defibrinated sheep blood. The resulting medium was mixed and poured into petri dishes.
Culture and microscopy
The laboratory identification of S. pneumoniae isolates relies on the recognition of typical morphological characteristics and the results of a few phenotypic tests. During microscopic evaluation, S. pneumoniae appears as lancet-shaped, Gram-positive diplococci or chains of cocci.3 Each blood sample was inoculated onto Blood Agar, MacConkey agar, TSSBA, and TSSBA-G media. Cultures were incubated at 35-37 °C in 5% CO‚ for 24-48 hours. S. pneumoniae was identified by colony morphology and Gram staining, appearing as lancet-shaped, Gram-positive diplococci or chains.
Optochin susceptibility test
Additional confirmation of S. pneumoniae was performed using the optochin susceptibility test. A 5 µg optochin disk (6 mm) was placed on a lawn culture of the isolate grown on 5% sheep blood agar. The plates were incubated at 35-37 °C in 5% CO2 for 18-24 hours. S. pneumoniae was identified by the presence of a zone of inhibition measuring ≥14 mm around the optochin disk, whereas viridans streptococci exhibited resistance with a smaller or no zone of inhibition.
Identification and antimicrobial susceptibility testing
The identification was started by colony microscopy with Gram staining. Colonies grown on both routine and selective media (TSSBA-G and TSSBA) were identified using standard laboratory procedures. Antimicrobial susceptibility testing was then performed with the Vitek 2 system using GP and AST-GP67 cards (bioMerieux), and results were interpreted based on Clinical and Laboratory Standards Institute (CLSI) breakpoint criteria.20
Data analysis
The proportion of positive cultures for each medium was calculated and presented as percentages and bar plots. The McNemar test was used to assess the statistical significance of differences between media types with a significance threshold of α = 0.05 (95% confidence) applied, and p-values <0.05 were considered statistically significant. Analyses were performed in R version 4.5.0, and additional visualization was performed in Python version 3.11.7.
Out of 45 blood samples from patients suspected of pneumonia and bacteremia, various bacterial growth patterns were observed across all tested culture media, as summarized in Table 1. Using Tryptic Soy Sheep Blood Agar with Gentamicin (TSSBA-G) and incubation under anaerobic conditions with 5% CO₂, Streptococcus pneumoniae was successfully isolated from 6 samples (13%), as shown in Table 2. In contrast, 11 samples (24%) demonstrated bacterial growth on TSSBA without gentamicin; however, not all isolates were identified as S. pneumoniae. The comparative performance and detection efficiency of each culture medium, including their ability to recover S. pneumoniae, are summarized in Table 3.
Table (1):
Bacterial Growth from Blood Samples Using Different Culture Media (n = 45)
| Media Type | Total Positive Cultures n (%) | Total Negative Cultures n (%) | non-S. pneumoniae Identified Bacteria (n) |
|---|---|---|---|
| Routine Media | |||
| Blood Agar | 12 (27%) | 33 (73%) | S. aureus (3), S. haemolyticus (1), K. pneumoniae (1), A. baumannii (1), P. fluorescens (1), B. cepacia (1), P. mirabilis (1), Pseudomonas sp. (1), Micrococcus luteus (1), S. epidermidis (1) |
| MacConkey Agar | 6 (13%) | 39 (87%) | *Only Gram-negatives bacilli 6 (13%) |
| Selective Media | |||
| TSSBA-G | 6 (13%) | 39 (87%) | – |
| TSSBA (without
gentamicin) |
11 (24%) | 34 (76%) | S. aureus (3), Micrococcus luteus (1), S. epidermidis (1) |
Table (2):
Detection of Streptococcus pneumoniae Across Different Culture Media (n = 45)
| Media Type | S. pneumoniae detected n (%) |
|---|---|
| Routine Media | |
| Blood Agar | 0 (0%) |
| MacConkey Agar | 0 (0%) |
| Selective Media | |
| TSSBA-G | 6 (13%) |
| TSSBA (without gentamicin) | 6 (13%) |
Table (3):
Total Bacterial Growth on Routine and Selective Media
| Media Type | Total Positive Cultures n (%) | S. pneumoniae detected n (%) |
|---|---|---|
| Routine Media | ||
| Blood Agar | 12 (27%) | 0 (0%) |
| MacConkey Agar | 6 (13%) | 0 (0%) |
| Selective Media | ||
| TSSBA-G & TSSBA (without gentamicin) | 11 (24%) | 6 (13%) |
| Total Bacterial Growth | 29 (64%) | |
Recent studies emphasize that media formulation, incubation atmosphere (e.g., 5% CO2), and enriched nutrient content significantly enhance S. pneumonia viability and detection.21,22 Additionally, antibiotic administration before sample collection has been shown to reduce culture sensitivity, leading to false negatives and underreporting.23
TSSBA yielded 11 positive bacterial cultures (24%), but these included mixed flora such as Micrococcus luteus and Staphylococcus epidermidis, organisms often considered contaminants in blood cultures. In contrast, isolates grown on TSSBA-G exhibited the characteristic colony morphology of S. pneumoniae. Notably, while routine blood agar yielded 12 positive cultures (27%), none of these isolates were identified as S. pneumoniae. (Table 2).
The use of tryptic soy sheep blood agar, particularly when supplemented with gentamicin, significantly increased the yield of positive blood cultures, including specific recovery of S. pneumoniae. The tryptic soy agar (TSA) media is recommended for use as a general-purpose growth medium for the isolation, cultivation, and differentiation of a wide variety of microorganisms. The addition of 5% sheep blood from healthy and antibiotic-free donor animals. This media can also be used as a growth medium for other organisms than bacteria, namely yeast and fungi. The selective use of gentamicin in TSSBA-G was intended to suppress the growth of non-target bacteria, thereby enhancing the recovery of S. pneumoniae compared to TSSBA without gentamicin. No additional enrichment media, such as Todd Hewitt Broth, were employed in this study. The macroscopic characteristics of bacterial colonies growing on TSSBA media without gentamycin were large and slimy, whereas those on TSSBA with gentamycin were small.
Previous work demonstrates that gentamicin-supplemented blood agar increases yield and suppresses competing organism, thus improving detection sensitivity.13 Such selective media have been validated in multiple settings for enhancing recovery from polymicrobial or contaminated specimens.17 The primary culture results on TSSBA-G media incubated in a CO2 incubator can be seen in Figure 1.
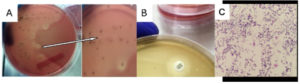
Figure 1. (A) S. pneumoniae Growth on TSSBA-G; (B) Optochin Disc Diffusion Test showing zone of inhibition confirming S. pneumoniae. (C) Microscopic appearance of S. pneumoniae
Figure 1 (A) shows that S. pneumoniae colonies on TSSBA-G α-hemolytic media appear within <24 hours, but the center of the pneumococcal colony becomes depressed after 24-48 hours. Figure 1 (B) shows a clear zone around an optochin disc (ethylhydrocupreine), indicating S. pneumoniae sensitivity. The TSSBA-G media supports the growth of pure S. pneumoniae, while TSSBA without gentamicin shows contamination by Micrococcus luteus and Staphylococcus epidermidis. As shown in Figure 1C, colonies grown on TSSBA-G displayed characteristic morphology consistent with S. pneumoniae.
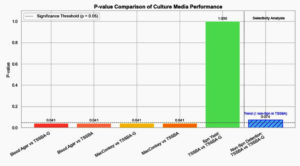
These findings are further supported by Figure 2, which presents the p-values from McNemar’s test comparing media performance. TSSBA-G significantly outperformed Blood Agar and MacConkey Agar (p = 0.041), while no significant difference was observed between TSSBA and TSSBA-G in isolating S. pneumoniae (p = 1.000). However, gentamicin appeared to significantly reduce the presence of non-S. pneumoniae organisms (p = 0.073), indicating its value in reducing contamination.
Contamination in blood cultures can result from several sources, predominantly due to improper skin disinfection during sampling.24,25 The use of selective culture media, particularly those containing antimicrobial-binding agents, has proven effective in minimizing false positives caused by skin flora such as Micrococcus spp. and coagulase-negative staphylococci (CoNS).26,27 Studies on blood culture stewardship and predictive models indicate that improved protocols (sample collection, media selection, and handling) can reduce contamination and increase pathogen yield, supporting the operational feasibility of selective media such as TSSBA-G.16,28,29
More than a third of septic patients had positive blood cultures, primarily due to inadequate sampling volume (50% of adults had <1.0 CFU/mL of blood) and prior antibiotic use. Additionally, 20%-30% of septic patients receive empiric antibiotics, which can reduce the likelihood of positive blood cultures.13 This study demonstrates that the use of TSSBA-G media, incubated anaerobically in a CO2 incubator, can increase the rate of positive blood cultures.
Implementing selective media approaches like TSSBA-G holds promise especially in resource-limited settings where molecular diagnostics may not be accessible. Recent evidence suggests that combining selective media with faster subculture and incubation workflows can produce clinically meaningful improvements in detection without major increases in cost or complexity.
The use of Tryptic Soy Sheep Blood Agar with and without gentamicin (TSSBA-G and TSSBA) significantly increased the blood culture positivity rate for S. pneumoniae, the leading cause of bacteremia and pneumonia. While routine media (Blood agar and MacConkey) yielded positive cultures in 27% of samples (12/45), none isolated S. pneumoniae. In contrast, TSSBA-G isolated S. pneumoniae in 6 out of 45 samples (13%), and TSSBA detected a broader range of organisms in 24% of samples (11/45), albeit with more contaminants. Statistical analysis using McNemar’s test confirmed that both TSSBA-G and TSSBA detected significantly more pathogens than routine media (p = 0.041).
This study highlights the unique application of TSSBA agar, particularly the modified formulation with gentamicin (TSSBA-G), for the selective isolation of S. pneumoniae from blood culture specimens. The primary advantage of this approach lies in the ability of gentamicin supplementation to suppress competing flora, thereby improving the recovery of S. pneumoniae, which is often overgrown or masked in conventional blood agar cultures. From a long-term perspective, this method offers the potential to enhance routine laboratory detection of S. pneumoniae in resource-limited settings, where access to automated systems may be restricted. The simplicity, cost-effectiveness, and adaptability of TSSBA-G make it a promising alternative for broader clinical use, especially for surveillance programs and diagnostic workflows in hospitals with high burdens of pneumonia and bacteremia. Furthermore, the adoption of such selective media could support more accurate epidemiological studies and antimicrobial resistance monitoring over time.
ACKNOWLEDGMENTS
The authors would like to thank the Research Institute of Universitas Sumatera Utara for providing funds, and the director, doctors, nurses, and clinical microbiology laboratory assistants at the Universitas Sumatera Utara Hospital, for contributing to the sample collection process.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the funds from TALENTA USU 2019 based on the scheme of Research Development of Universitas Sumatera Utara Hospital (PPRSU) with contract number: 496/UN5.2.3.1/PPM/KP-TALENTA USU/2019.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Health Research Ethical Committee of the Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia, vide reference number 334/TGL/KEPK FK USU-RSUP HAM/2019.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Chen H, Matsumoto H, Horita N, Hara Y, Kobayashi N, Kaneko T. Prognostic factors for mortality in invasive pneumococcal disease in adults: a systematic review and meta-analysis. Sci Rep. 2021;11(1):11865.
Crossref - Purba AK, Ascobat P, Muchtar A, et al. Multidrug-Resistant Infections Among Hospitalized Adults With Community-Acquired Pneumonia In An Indonesian Tertiary Referral Hospital. Infect Drug Resist. 2019;12:3663-3675.
Crossref - Tian PP, Su SS, Zhu LS, et al. Short-term culture for rapid identification by mass spectrometry and automated antimicrobial susceptibility testing from positive bottles. BMC Infect Dis. 2024;24(1):566.
Crossref - Lee YH, Kang MS, Yang J, et al. Comparison of conventional culture and automated blood culture system for microbiologic diagnosis of pleural infection. Microbiol Spectr. 2025;13:e00352-25.
Crossref - World Health Organization. Global Immunization Vision and Strategy 2006-2015. Switzerland: WHO; 2005. https://s3.amazonaws.com/wp-agility2/measles/wp-content/uploads/2017/01/Global-Immu-Strategy-2006-2015.pdf
- Reller LB, Weinstein MP, Werno AM, Murdoch DR. Laboratory diagnosis of invasive pneumococcal disease. Clin Infect Dis. 2008;46(6):926-932.
Crossref - Roberts T, Chandna A, Watthanaworawit W, et al. Impact of delayed processing of positive blood cultures on organism detection: a prospective multi-centre study. BMC Infect Dis. 2022;22(1):517.
Crossref - Harris AM, Bramley AM, Jain S, et al. Influence of antibiotics on the detection of bacteria by culture-based and culture-independent diagnostic tests in patients hospitalized with community-acquired pneumonia. Open Forum Infect Dis. 2017;4(1):ofx014,.
Crossref - Cobo F, Cabezas-Fernández MT, Cabeza-Barrera MI. Streptococcus pneumoniae bacteremia: clinical and microbiological epidemiology in a health area of Southern Spain. Infect Dis Rep. 2012;4(2):e29.
Crossref - Jorge EV, Meagan NW, Uriel AZ, et al. Prophylactic inhibition of colonization by Streptococcus pneumoniae with the secondary bile acid metabolite deoxycholic acid. 2021;05.17.444594.
Crossref - Iglesies-Torrent J, Garrido M, Morales J, Pitart C, Vergara A, Fernández-Pittol M. Enhancing pneumococcal bacteraemia diagnosis: A comparative assessment of culture-independent assays (MALDI-TOF-MS Sepsityper® module and a lateral flow inmunochromatography test). Enferm Infecc Microbiol Clin (Engl Ed). 2024;42(7):377-379.
Crossref - Blaschke AJ. Interpreting Assays for the Detection of Streptococcus pneumonia. Clin Infect Dis. 2011;52(4):331-337.
Crossref - Schmid RE, Washington JA 2nd, Anhalt JP. Gentamicin-blood agar for isolation of Streptococcus pneumoniae from respiratory secretions. J Clin Microbiol. 1978;7(5):426-427.
Crossref - Russell FM, Biribo SSN, Selvaraj G, et al. As a Bacterial Culture Medium, Citrated Sheep Blood Agar Is a Practical Alternative to Citrated Human Blood Agar in Laboratories of Developing Countries. J Clin Microbiol. 2006;44(9):3346-3351.
Crossref - World Health Organization. Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. 2003. https://arsp.com.ph/wp-content/uploads/2019/02/Manual-for-Lab-ID-AST-cdc-who.pdf
- Samuel L. Direct-from-Blood Detection of Pathogens: a Review of Technology and Challenges. J Clin Microbiol. 2023;61:e00231-21.
Crossref - Chen C, Fang J, Li X, et al. Diagnostic accuracy of non-culture methods for the diagnosis of melioidosis: a systematic review and meta-analysis. BMC Infect Dis. 2025;25(1):851.
Crossref - Saha S, Darmstadt G, Naheed A, et al. Improving the sensitivity of blood culture for Streptococcus pneumoniae. J Trop Pediatr. 2011;57(3):192-196.
Crossref - Arbefeville SS, Timbrook TT, Garner CD. Evolving strategies in microbe identification- a comprehensive review of biochemical, MALDI-TOF MS and molecular testing methods. J Antimicrob Chemother. 2024;79(12 Suppl 1):i2-i8.
Crossref - Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 30th ed CLSI supplement M100. Clinical and Laboratory Standards Institute. USA. 2020. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf
- Miellet WR, Almeida ST, Trzciński K, Sá-Leão R. Streptococcus pneumoniae carriage studies in adults: Importance, challenges, and key issues to consider when using quantitative PCR-based approaches. Front. Microbiol. 2023;14:1122276
Crossref - Muktiarti D, Khoeri MM, Tafroji W, Waslia L, Safari D. Serotypes and antibiotic susceptibility profile of Streptococcus pneumoniae isolated from nasopharynges of children infected with HIV in Jakarta, Indonesia, pre- and post-pneumococcal vaccination. Access Microbiol. 2021;3(3):000215.
Crossref - Elnoby AS, Nahla R. The Clinical Utility and Stewardship Challenges of Time to Blood Culture Positivity. Hosp Pharm. 2025;00185787251364488.
Crossref - Weinstein MP. Blood culture contamination: persisting problems and partial progress. J Clin Microbiol. 2003;41(6):2275–2278.
Crossref - Christenson RH, Snyder SR, Shaw CS, et al. Laboratory medicine best practices: systematic evidence review and evaluation methods for quality improvement. Clin Chem. 2011;57(6):964-969.
Crossref - Doern GV, Carroll KC, Diekema DJ, et al. Practical Guidance for Clinical Microbiology Laboratories: A Comprehensive Update on the Problem of Blood Culture Contamination and a Discussion of Methods for Addressing the Problem. Clin Microbiol Rev. 2019;33(1):e00009-19.
Crossref - Petit H, de Tymowski C, Dudoignon E, et al. Epidemiology and Outcomes of Bloodstream Infections in Patients in a Burns Intensive Care Unit: An 8-Year Retrospective Study. Open Forum Infectious Diseases. 2025;12(4).
Crossref - Ak K, Arabaci C. Resistance of Streptococcus pneumoniae Strains Isolated from Clinical Samples to Various Antibiotics. Dicle Med J. 2020;47(4):779-786.
Crossref - Townsa ML, Jarvisb WR, Hsueh PR. Guidelines on Blood Cultures. J Microbiol Immunol Infect. 2010;43(4):347-349.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.