ISSN: 0973-7510
E-ISSN: 2581-690X
Since rhizobacterial strains play vital role in plant growth, objective of this research was to isolate antagonistic rhizobacterial strains namely, B. cepacia, C. freundii and S. marcescens from mustard plant rhizospheric soil sample based on morphological, biochemical and molecular identification. The16S rRNA gene sequences of rhizobacterial isolates Burkholderia cepacia, Citrobacter freundii and Serratia marcescens were submitted to GenBank with accession numbers LC169488, LC169489, LC169490 respectively. Some biochemical test were performed like siderophore production test, HCN test, cellulase and lipase test to estimate different mechanisms adopted by our isolates to inhibit fungal and bacterial phytopathogens. The rhizobacterial isolates were investigated for the ability to perform as antagonistic biocontrol control agents to inhibit fungal pathogens namely Rhizoctonia solani and Phytophthora infestans that causes stem canker, black scurf in potato and late blight or potato blight disease respectively. The rhizobacterial isolates were also screened for their ability to inhibit Staphylococcus aureus and Escherichia coli that are common bacterial pathogens. Antibiotic susceptibility profile of our isolates was also observed. Present study thus reveal that our isolated strains B. cepacia, C. freundii and S. marcescens are efficient biocontrol agents and can be used as potential biofertilizers to wean our society off of its widespread use to agricultural chemicals.
Rhizobacteria, antifungal activity, antibiotic susceptibility.
Quality and quantity of food are going to be important challenges to overcome in coming future. Exceeding population growth rate requires production of more agricultural products and to inevitably move towards increased production per unit area. Since fertilizer management is considered as one of the main factors of sustainable agriculture, gradual replacement of chemical fertilizers with biological fertilizers is quite inevitable since biofertilizers are environment friendly and cost-effective. Elucidating non-chemical control methods to reduce postharvest decay is becoming increasingly important (Conway et al. 2004). Naturally occurring antagonists on host surfaces are a promising component of biological crop protection (De Costa and Erabadupitiya 2005). The rhizosphere, the narrow zone of soil that surrounds and influences the plant roots, is home to a large number of microorganisms and is considered to be one that can have profound effects on the growth, nutrition and health of plants in agro-ecosystems (Berendsen et al. 2012). Bacteria able to colonize plant root systems and promote plant growth are referred to as plant growth promoting rhizobacteria (PGPR) (Kloepper and Schroth 1978). PGPR can affect plant growth by a wide range of mechanisms such as solubilization of inorganic phosphate, production of phytohormones, siderophore and organic acids, lowering of plant ethylene levels, nitrogen fixation and biocontrol of plant diseases (Rani et al. 2012). Along with the promotion of plant growth, PGPR also help in sustainable agricultural development, thereby protecting the environment and thus help in maintaining the ecological balance. Therefore, the use of such beneficial bacteria as biofertilizers and biocontrol agents has attracted increased interest worldwide in attempts to achieve sustainability in agriculture (Datta et al. 2011).
The Biocontrol bacterial species generally employ an array of mechanisms such as antibiosis, competition, production of hydrocyanic acid (HCN), siderophore and antifungal compounds to antagonize pathogens (Singh et al. 2010). Various rhizobacteria, including for example Burkholderia cepacia, Staphylococcus epidermidis, and strains of the Bacillus subtilis group, stimulate plant growth by the emission of volatile organic compounds such as hydrogen cyanide and ammonia (Bitas et al. 2013). Voisard et al. (1989) reported that HCN production by the rhizobacteria plays a significant role in the biological control of pathogens by acting as an inducer of plant resistance. It was also reported that HCN indirectly influences plant growth promotion (Kumar et al. 2012). Afsharmanesh et al. (2010) suggested that fungal growth is mainly inhibited by HCN production and siderophore production. This in turn can indirectly enhance the plant growth by keeping plant healthy and disease free (Glick and Pasternak 2003). Additionally, secretion of siderophore and production of chitinases, glucanases, cellulases, lipases and other lytic enzymes by PGPR can affect plant growth as these are also found to suppress the growth of fungal pathogens (Kloepper et al. 1980). Siderophores can be defined as small peptidic molecules containing side chains and functional groups that can provide a high-affinity set of ligands to coordinate ferric ions (Crosa and Walsh 2002). Siderophores are secreted to solubilize iron from their surrounding environments, forming a complex ferric- siderophore that can move by diffusion and be returned to the cell surface (Andrews et al. 2003). The potent siderophore, pyoverdin, for example, can inhibit the growth of bacteria and fungi that present less potent siderophores in iron-depleted media in vitro (Kloepper et al. 1980a). A pseudobactin siderophore produced by P. putida B10 strain was also able to suppress Fusarium oxysporum in soil deficient in iron; this suppression was lost when the soil was replenished with iron, a condition that represses the production of iron chelators by microorganisms (Kloepper et al. 1980b). Burkholderia cepacia has shown antagonistic activity against a broad range of plant pathogens (Cartwright and Benson 1995). Several studies suggested that pyrrolnitrin production by B. cepacia as well as other Pseudomonas spp, is closely associated with biocontrol of plant diseases (Hwang et al. 2002). Serratia species, usually found in diverse natural environments, have been found to reduce disease severity of various foliar and soilborne diseases. Among Serratia species, S. marcescens was reported as an important bacterium that has the ability to induce systemic resistance to various pathogens and to exhibit antifungal activity against several soil-borne fungi like Rizoctonia solani and Phytophthora infestans that causes stem canker, black scurf in potato and late blight or potato blight disease respectively.
The disease protection measures of medicinal plants are still restricted to the application of various chemical fungicides which strictly do not fit with the basic theory of usefulness of herbal drugs. Also, the residual effects of different chemicals eventually contaminate the purity of such plant drugs and are also of serious concern from environmental point of view (Sharma et al. 2004). Many researchers have reported that effective colonization by PGPR contributed to the successful suppression of plant pathogens and thus these can be considered as potential use of biocontrol agents as replacements for agrochemicals (Guo et al. 2007). In recent years, certain plant associated rhizobacteria belonging to various genera like Pseudomonas, Burkholderia and Bacillus have drawn attention because of their potential to suppress the soil borne pathogens causing diseases in different crops (Cazorla et al. 2007). Due to excessive use of various agrochemicals including various pesticides of different chemical families in crop production, these beneficial microorganisms and their physiological activities important to soil fertility are adversely affected (Srinivas et al. 2008). Bacteria strains isolated from soils that have been contaminated with various biochemicals, including pesticides, are increasingly being used for bioremediation (reclaiming the soil by inoculating it with organisms able to degrade certain compounds which are detrimental to other life forms). In other words, the bacteria are using specific pesticides to meet their energy needs, i.e., using them as food (Hernández et al. 2008). To overcome the deleterious effects of pesticides on plants and of soil fumigants on soil fertility, the treatment of seeds with PGPR could be used as a bio-inoculant which displays a wide range of tolerance to pesticides and exhibit PGP activities under pesticide-stress (Ahemad and Khan 2010).
Isolation of antagonistic rhizobacterial strains
When Indian mustard plant was found, a 6″X 2.5″X 5″ soil sample was collected, the soil was serially diluted and appropriate dilutions (10-3, 10-5, 10-7) were spread plated on nutrient agar medium. The plates were incubated at 38°C and developed colonies were identified on the basis of cultural, morphological characteristics as described in the Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994). Selective colonies were taken and streaked on selective media for individual strains and kept in incubator at 38°C for 24 h to obtain colonies.
Identification of specific bacterial species
The bacterial colonies on incubated plates were identified on the basis of cultural, morphological and biochemical characteristics as described in the Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994).
Cultural and Morphological Characteristics
The isolates were identified on the basis of different colony characteristics like diameter, consistency, colour, texture, elevation, margin etc. The organism was subjected to Gram’s staining (Holt et al. 1994) for observing the morphological characteristics such as the shape and arrangement (clusters or chains) of cells, and gram-reaction.
Screening of Rhizobacteria for plant growth promoting traits
Siderophore production
For siderophore production, 1µl of overnight raised rhizobacterial culture in LB broth were spotted on Chrome Azurol S (CAS) agar plates and incubated at 37°C for 48 h. Plates were observed for the appearance of orange halo around the bacterial colony (Ali et al. 2013).
HCN Production
For qualitative estimation of HCN, isolate was streaked on nutrient agar plate supplemented with 4% glycine. A whatman filter paper soaked in a solution of 2% Na2CO3 and 0.5% picric acid was placed between base and lid of petriplate and incubated at 37°C in inverted position for 48 h and observed for color change from yellow to orange brown (Ali et al. 2013).
Chitinase test
For the Chitinase test spot inoculation of isolated strain was made on chitin agar plate amended with 2% phenol red and incubated for 48 h at 37°C. Presence of clear zone around the culture indicates the chitinase activity (Holt et al. 1994).
Protease production
The bacterial species were spot inoculated on nutrient agar containing casein and gelatin. The zone of hydrolysis was observed for the protease production (Pandey et al. 2013).
Lipase test
Lipolytic activity was carried out on trimethoprim plate and observation of halogen was considered as positive for lipase production.
Antibiotic sensitivity test
Isolate bacterial strains were tested for its resistance against standard antibiotics namely gentamicin (30 µg), ampicillin (10 µg), erythomycin (10 µg), kanamycin (5 µg), tetracyclin (10 µg), vancomycin (25 µg), and chloramphenicol (10 µg) by the antibiotic sensitivity assay. Briefly, the bacterial cultures were swabbed onto NA media plates. The standard antibiotic disc (6 mm) was placed over the media surface and the plates were incubated at 37 °C for 24 h. The experiment was done in triplicate. The results were interpreted on the basis of the diameter of inhibition zone using the zone size
Antagonistic test
Biocontrol ability of the isolate was evaluated by fungal mycelial inhibition by well diffusion method against the bacterial (E. coli and S. aureus) and fungal (R. solani and P. infestans) pathogens. After the solidification of potato dextrose agar (PDA) 100µl of fungal spore suspension in 0.85% sterile saline was spread on the solidified plate. Well diameter of 6mm was made by metallic borer and filled with overnight grown culture of isolate (1 × 108cfu) and kept for incubation at 28°C for seven days.
Bacterial DNA Isolation
DNA was isolated using Phenol Chloroform Method. From the overnight grown liquid culture 1.5 ml of culture was taken in micro centrifuge tube and centrifuged at 8000 rpm for 5 mins and pellet the bacterial cells. Pellets were suspended in 900µl of the TE buffer. 1/10 volume of 10% SDS solution (900µl solution then add 100µl of solution into it) was then added and tubes were kept at 50-60ºC under water bath for approximately 2 hours. Mix 600µl of Phenol:Chloroform:Isoamyl alcohol(25:24:1) to it and shake gently by repeatedly inverting the tube. White precipitate appeared after mixing, this mixture was added to the tube and solution became creamy orange (white due to protein precipitation and orange due to phenol). Kept for 5 min and then centrifuged at 10000 rpm for 10 min. Upon centrifugation 3 layers were appeared. Upper transparent aqueous layer was collected in fresh new eppendorf tube and discard the lower layer. Kept the eppendorf tube in refrigerator to cool it for 10-15 min. Double volume of chilled propanol was added to the aqueous DNA solution drop by drop from its wall. After adding DNA precipitate out from the solution and which was kept again on 0ºC for 15 min. After precipitation, tubes were centrifuged at 10000rpm for 10 min and supernatant was discarded. DNA pellet was dried in air and dissolved in 50µl TE buffer and then run on gel with 1kb ladder (Ali et al. 2013).
16S rRNA amplification
For molecular characterization, bacterial genomic DNA was quantified at 260/280nm. The 16SrRNA gene was amplified by PCR using forward 5′-CCGAATTCGTCGACAACAG AGTTTGATCCTGGCTCAG-3′ and reverse primer 5′-CCCGGGATCCAAGCTTACG GCTACCTTGTTACGACTT-3 primers under standard conditions (initial denaturation 94°C for 3 min, 35 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30s, extension at 72°C for 60s, and final extension at 72°C for 7 min). The PCR product was found the size of 1424, 1487 and 1306 bp for Burkholderiacepacia, Citrobacter feurendii and Serratia marcescens respectively and was purified and sequenced (Applied Biosystems, New Delhi). The sequence of 16S rRNA genes of all three isolates were obtained and compared with the existing database of 16S rRNA gene and submitted to Gene Bank of NCBI.
Isolation of rhizobacterial strains from soil samples
In the present study three rhizobacterial strains (i.e. Burkholderia cepacia, Citrobacter freundii, Serratia marcescens) were isolated by spread plate technique on selective media from 10-3 dilution of rhizospheric soil of mustard plant. Mustard plant rhizospheric soil was also used as sample for isolation of PGPR by (Lugtenberg and Kamilova 2009). Hiltner (1904) discovered that the rhizospheric soil is abundant in bacterial population as compared to the rest bulk soil.
Identification of rhizobacterial strains from soil samples
Colonies which were identified morphologically as Burkholderia cepacia, Citrobacter freundii and Serratia marcescens respectively were streaked again on selective media to obtain pure culture and then performed gram staining. The morphological characteristics were recorded (Table 1). Previous study reported that PGPR isolated from Indian mustard rhizospheric soil, possessed ACC deaminase enzyme and promoted root proliferation significantly (Belimov et al. 2001). Dubey and Maheshwari (2010) isolated genera of Azatobacter, Bacillus, Burkholderia from soil which supply nitrogen to legume plants.
Table (1):
Cultural, morphological and biochemical characteristics of B. cepacia, C. freundii and S. marcescens isolate.
| Cultural characteristics | Colour of colony | opaque light yellow colonies | Light gray translucent to opaque colonies with glossy surface | Pink to magenta opaque iridescent colonies |
|---|---|---|---|---|
| Morphological characteristics | Shape of colony | Staright rods with rounded ends | Straight rods occurring singly or in pairs | Straight rods with rounded ends |
| Elevation | convex | convex | convex | |
| Gram’s reaction | Gram negative | Gram negative | Gram negative | |
| Spore formation | Non-spore forming | Non-spore forming | Non-spore forming | |
| Arrangement of cells | Cells occur singly, rarely in pairs | Cells form clusters | Cells form clusters | |
| Biochemical | Indole test | Negative | Negative | Negative |
| Methyl red test | Negative | Positive | Positive | |
| VP test | Negative | Negative | Positive | |
| Catalase test | Positive | Positive | Positive | |
| Citrate utilization test | Negative | Positive | Positive | |
| Nitrate reduction test | Positive | Positive | Positive | |
| Hydrogen sulphide test | Positive | Positive | Negative | |
| Oxidase test | Negative | Negative | Negative | |
| Urease test | Positive | Positive | Negative | |
| Starch hydrolysis | Negative | Negative | Negative | |
| ONPG | Positive | Positive | Positive | |
| PPA | Positive | Positive | Positive | |
| Carbohydrate Fermentation | D-Glucose | Positive | Positive | Positive |
| Sucrose | Positive | Positive | Positive | |
| Maltose | Positive | Positive | Positive | |
| Lactose | Negative | Positive | Negative | |
| Xylose | Positive | Positive | Negative | |
| D-Mannitol | Negative | Positive | Positive | |
| Raffinose | Positive | Positive | Positive | |
| L-Rhamnose | Positive | Positive | Negative | |
| L-Arabinose | Positive | Positive | Negative | |
| Esculin | Negative | Positive | Positive | |
| L-Arginine | Negative | Positive | Negative | |
| L-Ornithine | Positive | Positive | Positive | |
| L-Lysine | Negative | Negative | Positive |
Screening of Rhizobacteria for plant growth promoting traits
Screening of bacterial isolates for their ability to promote plant growth. Rhizobacteria exhibit several plant growth promoting characteristics like phosphate solubilization, Nitrogen fixation, production of siderophore, IAA, Ninhydrin and ACC deaminase (Fig.1,2,3) that was supported by study of Bhattacharyya and Jha, 2012 on B. cepacia, S. marcescens isolates. The bacteria included in PGPR category are distributed across different taxa comprising of Firmicutes, Acinetobacter, Cyanobacteria, Bacteriodes and Proteobacteria (Tilak et al. 2005). The proteobacteria falls in the genera Bacillus, Azatobacter, Arthrobacter, Klebsiella, Azospirillum, Enterobacter, Serratia, Burkholderia, Pseudomonas, Alcaligenes, (Somasegara and Hoben 1994).
Indole acetic acid production test
Indole acetic acid test was conducted (Fig. 1a) that showed 4.22 µg/ml concentration of Burkholderia cepacia, while lesser concentration values of 2.42 and 1.86 µg/ml (Fig.1a) were recorded for Citrobacter freundii, Serratia marcescens respectively. Results of Ali et al., 2013 showed 41-47 µg/ml concentration of IAA with species of Pseudomonas. In another research V. paradoxus 5C-2 was seen positive for indoles production in vitro (Jiang et al. 2012).
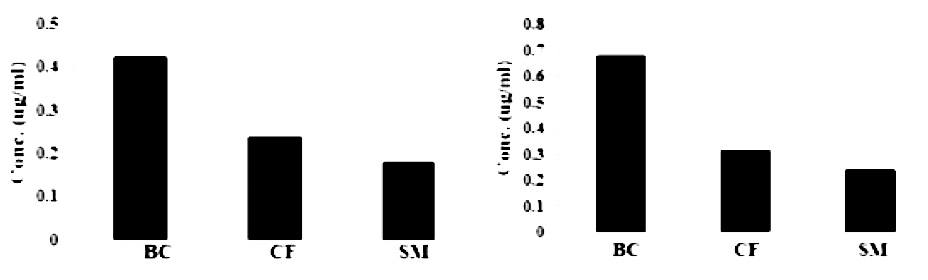 Fig. 1. Assay of (a) Indole acetic acid and (b) Gibberellic acid production for several rhizobia strains
Fig. 1. Assay of (a) Indole acetic acid and (b) Gibberellic acid production for several rhizobia strainsIAA production varies with bacterial isolates and concentration of tryptophan, although intrinsic IAA production ability of rhizospheric microorganism is regulated by the availability of precursors and uptake of rhizospheric IAA by plant (Arshad and Frankenberger 1993). (Belimov et al. 2009) reported that V. paradoxus 5C-2 strain elevated xylem abscisic acid concentrations in pea plant under drought conditions. Sharma et al. (2013) concluded in his study that all rhizobacterial isolates produced IAA, Bacillus produced (11.2-22.8 µg/ml) and Pseudomonas isolates (4.4-21.6 µg/ml).
Gibberellic acid production test
All three isolated bacterial strains were tested for gibberellic acid production where Burkholderia cepacia showed concentration i.e. 0.68µg/ml, while Serratia marcescens and Citrobacter freundii showed 0.24 and 0.32 µg/ml (Fig.1b) concentration respectively. From the graph (Fig.1b) it can be observed that Burkholderia cepacia showed more concentration compare to other two isolates. Our results support the concept that the growth promotion in plants induced by Azospirillum infection may occur by a combination of both gibberellin production and gibberellin glucoside or glucosyl ester de-conjugation by the bacterium (Piccoli et al. 1997). Tien et al. (1979) detected gibberellins-like substances in supernatants from A. brasilense cultures, at an estimated concentration of 0.05µg/ml GA3 equivalent. A quantitative estimation was done by the dwarf rice cv. Tan-ginbozu microdrop bioassay, showing that 20-40 pg/ml were produced (Bottini et al. 1989). The same gibberellins were found in similar amounts in cultures of A. brasilense (Janzen et al. 1992). The GAs pools in roots and nodules were of similar size, indicating that Rhizobium does not make a major contribution to GAs content in infected tissues (Atzorn et al. 1988).
ACC determination by colorimetric Ninhydrin assay
Ethylene glycol is used as solvent stabilizing ninhydrin reagent and for colour development. Ascorbic acid addition into ninhydrin reagent prevents oxidation of hydrindantin. Values for absorbance of ACC solutions were found in the rage 0.015 to 0.3 mmol/1 at 570 nm after the reaction was highly correlated with ACC concentrations (R2 = 0.999) and resulted in linear calibration curves by standard ninhydrin assay. Conc. of 0.05, 0.15 and 0.25 mmol/l was recorded for S. marcescens, C. freundii and B. cepacia respectively. The absorbance value of each ACC working concentration at wavelength of 570 nm in the standard assay was related to previous results of B. cepacia (Li et al., 2011) (Fig.2a). Research of Li et al. 2011 adds more functionality to the ninhydrin assay, although it does not accurately determine PGPR ACC deaminase activity, but provides a screening method with a relatively short turnaround time. Determination of ACC concentrations in rhizobacterial isolate after certain incubation period in the DF-ACC medium, by both assays were statistically identical at the 0.05 confidence level.

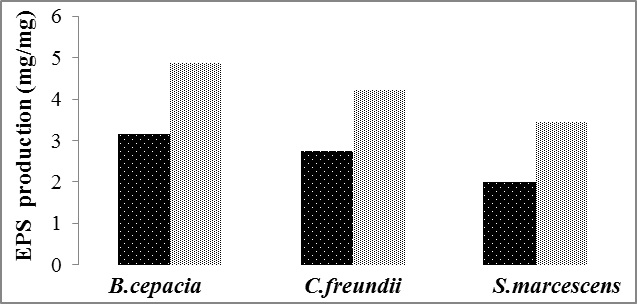 Fig. 2. Comparison of ACC Deaminase conc. for (a) Ninhydrin assay and (b) Exoploysaccharide production test of isolates
Fig. 2. Comparison of ACC Deaminase conc. for (a) Ninhydrin assay and (b) Exoploysaccharide production test of isolatesExopolysaccharide production test
Isolates screened for EPS production under both no stressed conditions as well as under minimum water potential (“0.30 MPa). The strain B.cepacia produced maximum amount of EPS (3.18 ± 0.02 mg/mg protein) under non-stressed condition, while and S.marcescens produced lesser amount of EPS (2.76 ± 0.04 and 2.01± 0.3 mg/mg protein respectively). Under drought stress, B. cepacia was best isolate since produce (4.893±0.06 mg/mg protein) of EPS followed by C.feurendii (4.23 ± 0.03 mg/mg protein) and S.marcescens (3.46±0.05 mg/mg protein) (Fig.2b).
Ali et al. (2014) conducted study on Pseudomonas isolates and found maximum 3.22 mg/mg protein EPS production (from Rdgp10 strain) while another strain (BriP15) produced 2.18 mg/mg protein EPS. Hartel and Alexander (1986) observed a significant correlation between the amount of EPS produced by cowpea Bradyrhizobium strains and desiccation tolerance. Glick et al. (1998) established that PGPB that have ACC deaminase activity help plants to withstand stress (biotic or abiotic) by reducing the level of the stress hormone ethylene through the activity of enzyme ACC deaminase, which hydrolyzes ACC into á-ketobutyrate and ammonia instead of ethylene.
Antibiotic and Bio control efficiency of selected rhizobacterial isolates
Different antibiotics discs were used to check the efficiency of selected isolates (Fig. 3) and found resistant against all except two for C. freundii. Zone were measured against selected antibiotics and recorded as gentamicin (12, 12, 7mm), ampicillin (9, 13, 11mm), erythomycin (10, 10, 3mm), kanamycin (14, 9, 11mm), tetracyclin (8, 10, 0mm), vancomycin (7, 18, 13 mm), and chloramphenicol (11, 11, 0) for B. cepacia, S. marcascens and C. freundii respectively.
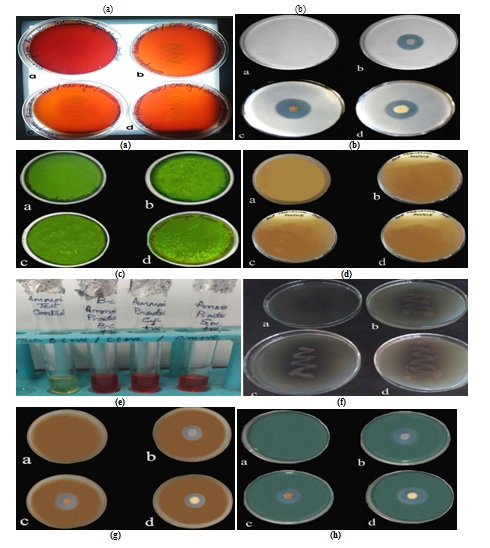 Fig. 3. Plant growth promoting (a) Siderophore production (b) Phosphate solubilisation (c) Nitrogen fixation (d) HCN production (e) Ammonia production (f) Cellulase production (g) Chitinase production and (h) Lipase production test for (a) Control (b) B. cepacia (c) S. marcescens (d) C. freundii
Fig. 3. Plant growth promoting (a) Siderophore production (b) Phosphate solubilisation (c) Nitrogen fixation (d) HCN production (e) Ammonia production (f) Cellulase production (g) Chitinase production and (h) Lipase production test for (a) Control (b) B. cepacia (c) S. marcescens (d) C. freundiiScreening of our rhizobacterial isolates was performed to estimate their efficacy to inhibit bacterial strains viz. E.coli (26, 25, 27mm) and S. aureus (27, 26, 13mm) while for fungal pathogen i.e. R. solani (17, 15, 14mm) and P. infestans (25, 14, 9mm) against B. cepacia, S. marcascens and C. freundii respectively (Fig.4-6).
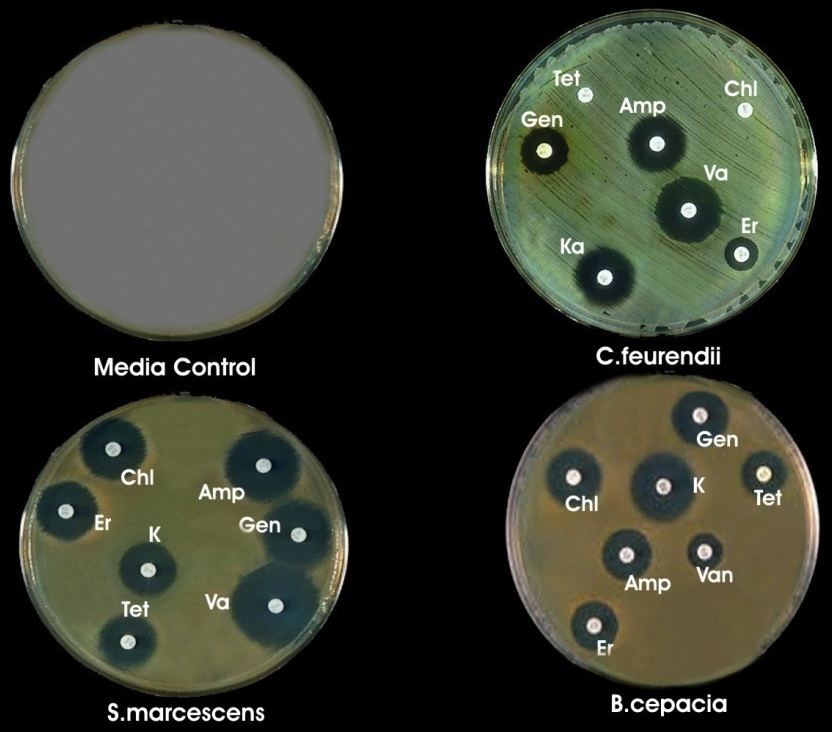 Fig. 4. Antibiotic sensitivity test (a) Control (b) C. freundii (c) S. marcescens (d) B. cepacia
Fig. 4. Antibiotic sensitivity test (a) Control (b) C. freundii (c) S. marcescens (d) B. cepacia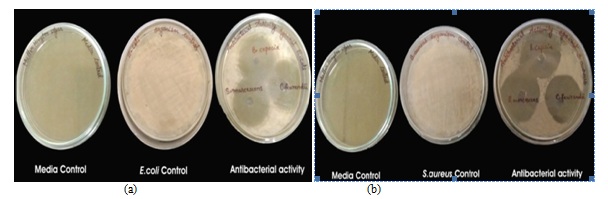 Fig. 5. Antibacterial efficiency of rhizobacterial isolates against (a) E. coli and (b) S. aureus
Fig. 5. Antibacterial efficiency of rhizobacterial isolates against (a) E. coli and (b) S. aureus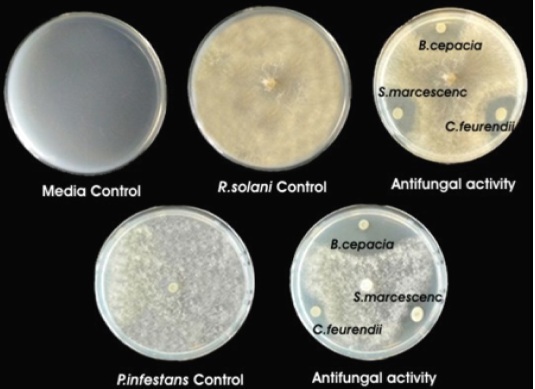 Fig. 6. Antifungal efficacy of rhizobacterial isolates against R. solani and P. infestans
Fig. 6. Antifungal efficacy of rhizobacterial isolates against R. solani and P. infestansIsolation and sequence characterization of Rhizobacterial 16S rRNA gene
Rhizobacterial genomic DNA was extracted and run on agarose gel with 1kb ladder. All three bacterial cultures showed genomic DNA size greater than 10kb size (Fig.7). Spectrophotometric analysis of samples was done to quantify rhizobacterial genomic DNA concentration and found 1.53 µg/ml for Burkholderia cepacia; 1.38 µg/ml for Serratia marcescens; 1.35 µg/ml for Citrobacter freundii . DNA from the above mentioned 03 Rhizobia strains were PCR amplified and found the size of 1424, 1487 and 1306 bp for Burkholderia cepacia; Citrobacter freundii; Serratia marcescens respectively (Fig.7). All three amplified products were then sequenced for 16S rRNA at Applied Biosystems using the oligonucleotide primers i.e. 16S forward primer: 5′-CCGAATTCGTCGACAA CAGAGTTTGATCCTGGCTCAG-3′ and 16S reverse primer: 5′-CCCGGGATCCAAGCTTACG GCTACCTTGTTACGACTT-3. After sequencing of all three bacterial isolates, sequences were submitted to DNA Database Bank of Japan (DDBJ), a collaborator of NCBI and obtained accession number i.e. LC169488 , LC169489 and LC169490 for Burkholderia cepacia; Serratia marcescens; Citrobacter freundii respectively for all three isolates. Analysis of these three sequences by comparison with the sequences of GenBank database indicated that 26 strains were Rhizobium leguminasarum with > 99% identities with the type strain (i.e. 1398/1402 bp).
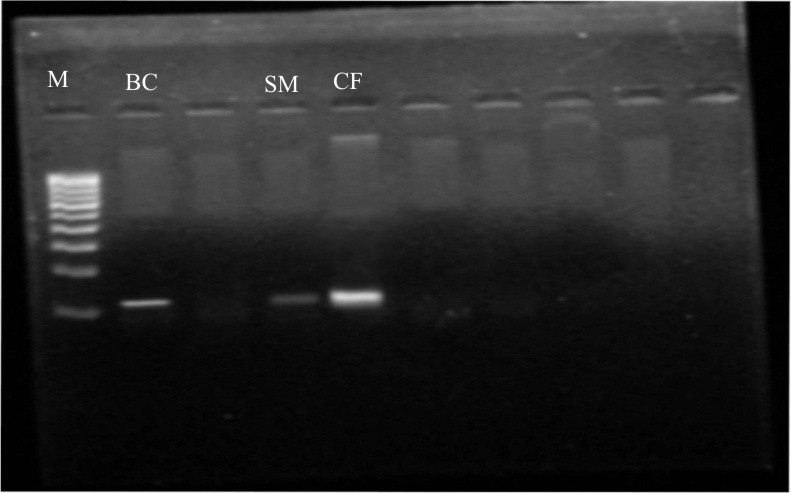 Fig. 7. PCR amplification for 16s rRNA gene of isolates (Lane 1: 1kb Ladder, Lane 2: B. cepacia, Lane 4: S. marcescens, Lane 5: C. freundii
Fig. 7. PCR amplification for 16s rRNA gene of isolates (Lane 1: 1kb Ladder, Lane 2: B. cepacia, Lane 4: S. marcescens, Lane 5: C. freundiiPGPR that exhibit ACC deaminase enzymatic activity support plants to survive under stress (biotic or abiotic) by lowering stress ethylene concentration by hydrolyzing ACC (immediate precursor of ethylene) into a-ketobutyrate and ammonia (Arshad et al. 2007). Variations in ACC deaminase enzymatic activity potential of the strains were noted in drought stress. In a previous research secretion of enzyme ACC deaminase by Pseudomonas strain SorgP4 was confirmed by acdS gene amplification using the same primer as reported earlier obtained a partial acdS gene from Pseudomonas isolate SorgP4 (Farajzadeh et al. 2010). acdS gene that codes for enzyme ACC deaminase has been earlier isolated from some rhizobacteria from rhizosphere of plant with or without abiotic stresses (Jha et al. 2009). Rhizobacteria capable of producing ACC deaminase are also known to posses potential to elevate growth of plants especially under stressed conditions such as flooding, heavy metals, high salinity and drought, thus acdS gene is a useful candidate gene for developing bio-inoculants to manage abiotic stress in plants. (Ali et al. 2013)
Bacterial 16S rDNA sequences are generally considered as highly conserved (Woese 1987). Evolutionary or taxonomic relationships among bacteria and rhizobacteria are usually estimated through 16S rDNA sequence comparisons. According to the 16S rRNA sequences of our three rhizobacteria strains which exhibit ACC deaminase enzymatic activity, they are identified to be Burkholderia cepacia; Serratia marcescens; Citrobacter freundii.
Similar sequences generally code for similar structure; similar structure are considered to be associated in similar function. Therefore BLAST2 was performed to evaluate the association of ribosomal genes and growth promotion (Table 2) among these three species. In contrast to our notion of similar sequence to similar function, the sequences were found relatively divergent. Therefore, to search for more similar homologs of individual sequences, BLASTN was performed with an output value of 10.
Table (2):
Homology based study for selected isolates.
Query value (%) Identity (%) |
B. cepacia |
C. freundii |
S. marcescens |
|---|---|---|---|
Burkholderia cepacia |
100 100 |
81 79 |
77 78 |
Citrobacter freundii |
77 79 |
100 100 |
84 91 |
Serratia marcescens |
85 78 |
96 91 |
100 100 |
Three rhizobacterial strains were isolated from soil sample of mustard plant rhizosphere. These isolates were subjected morphological and biochemical characterization followed by screened for plant growth promoting efficiency using several biochemical tests like phosphate solubilization, siderophore production, ammonia production, nitrogen fixation along with confirmatory tests like ninhydrin production, gibberellic acid production and indole acetic acid production tests. Our isolates were also screened for their ability to perform biocontrol activity using some antagonistic tests like HCN, chitinase, cellulase and lipase production tests. Genomic DNA of all three cultures were isolated and amplified through PCR with suitable primers. PCR products were then sequenced for 16s RNA gene and results were successfully submitted to DDBJ/NCBI under allotted accession numbers and bioinformatics based phylogenetic analysis was done to estimated 16s RNA sequence similarities of our isolates with those in the database and their evolutionary relationship. Based on molecular characterization selected three rhizobacterial strains were identified as Burkholderia cepacia, Serratia marcescens and Citrobacter freundii and these isolates were found capable for plant growth promoting activity and possessed significant antagonistic efficacy. Antibiotic susceptibility profile of B. cepacia, C. freundii and S. marcescens against various antibiotics namely gentamicin, ampicillin, erythomycin, kanamycin, tetracyclin, vancomycin and chloramphenicol was recorded. The ultimate achievement of these inoculates will be to be use as bio fertilizers that are cost effective and do not hinder soil fertility unlike chemical fertilizers. Further research and understanding of mechanism of PGPR mediated phyto-stimulation would pave the way to find out more competent rhizobacterial strains which may work under diverse agro-ecological conditions.
- Afsharmanesh, H., Ahmadzadeh, M., Javan-Nikkhah, M., Behboud, K. Characterization of the antagonistic activity of a new indigenous strain of Pseudomonas fluorescens isolated from onion rhizosphere. J. Plant Pathol., 2010; 92: 187-194.
- Ahemad, M., Khan, M. Effects of pesticides on plant growth promoting traits of Mesorhizobium strain MRC4. J. Saudi Soc. Agri. Sci., 2010; 11: 63-71.
- Ali, S., Shya, V., Rao, L.V. Isolation characterization of drought-tolerant ACC deaminase exopolysaccharide- producing fluorescent Pseudomonas sp. Ann. Microbiol., 2013; 5: 1-10.
- Andrews, S., Robinson, A.. Rodriguez-Quinones. F. Bacterial iron homeostasis. FEMS microbial. Rev. 2003; 27: 215-237.
- Arshad, M., Frankenberger, W.T. Ethylene: Agricultural Sources and Applications. Kluwer Academic/Plenum Publishers, New York. 2002. pp 137-138.
- Arshad, M., Saleem, M., Hussain, S. Perspectives of bacterial ACC-deaminase in phytoremediation. Trends Biotechnol., 2007; 25: 356-362.
- Atzorn, R., Crozier, A., Wheeler, C., Sberg, G. Production of gibberellins indole-3- acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Plant, 1998; 175: 532-538.
- Belimov, A., Dodd, I., Hontzeas, N., Theobald, J., Safronova V., Davies, W. Rhizosphere bacteria containing 1-aminocyclopropane- 1-carboxylate deaminase increase yield of plants grown in drying soil via both local systemic hormone signalling. New Phytol., 2009; 181: 413–423.
- Belimov, A., Safronova, V., Sergeyeva, T., Egorova, T., Matveyeva, V., Tsyganov, V., Borisov, A., Tikhonovich, I., Kluge, C., Preisfeld, A., Dietz, K., Stepanok, V. Characterization of plant growth promoting rhizobacteria isolated from polluted soils containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol., 2001; 47: 642-652.
- Berendsen, R, Pieterse, C., Bakker, P. The rhizosphere microbiome and plant health. Trends Plant. Sci., 2012; 17: 478–486.
- Bitas, V., Kim, H., Bennett, J., Kang, S. Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol. Plant Microbe. Interact. 2013; 26: 835–843.
- Bottini, R., Fulchieri, M., Pearce, D., Pharis, R. Identification of gibberellins A1 and A3 isolates A3 in cultures of Azospirillum lipoferum. Plant Physiol. 1989; 90: 45-47.
- Cartwright, D., Benson, D. Optimization of biological control of rhizoctonia stem rot of poinsettia by Paecilomyces lilacenus and Pseudomonas cepacia. Plant Dis. 1995; 79: 309-313.
- Cazorla, F., Romero, D., Pérez-García, A., Lugtenberg, B. de Vicente, A., Bloemberg, G. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol., 2007; 103: 1950-1959.
- Conway, C., Sulzman, C., Raulston, B. Factors affecting detection probability of California black rails. J. Wildlife Mgmt., 2004; 62: 360-370.
- Crosa, J. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev., 1989; 53: 517–530.
- Decosta, D., Erabadupitiya, H. An integrated method to control postharvest diseases of banana using a member of the Burkholderia cepacia complex. Posth. Biol. Technol. 2005; 36: 31–39.
- Dubey, R., Maheshari, D. A Text book of Microbogy. S. Ch, India. 2010. pp187.
- Dutta, S., Shome, A., Kar, T. Das, P. Counterion-Induced Modulation in the Antimicrobial Activity and Biocompatibility of Amphiphilic Hydrogelators: Influence of in-Situ-Synthesized Ag-Nanoparticle on the Bactericidal Property.Langmuir., 2011; 27: 5000-5008.
- Farajzadeh, D., Aliasgharzad, N., Bashir, N., Yakhchali, B. Cloning characterization of a plasmid encoded ACC deaminase form indigenous Pseudomonas fluorescens FY32. Curr. Microbiol., 2010; 61: 37-43
- Glick, B.R., Penrose, D., Li, J. A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol.,1998 190: 63–68.
- Glick, B.R., Pasternak, J.J. Molecular Biotechnology: Principles and Applications of Recombinant DNA. ASM Press. Michigan. 2003; pp 560
- Guo, Y., Zheng, H., Yang, Y., Wang, H. Characterization of Pseudomonas corrugate strain P94 isolated from soil in Beijing as a potential biocontrol agent, Curr. Microbiol., 2007; 55: 247-253.
- Hernandez, G., Tetweiler, G., Miron, M., Lasko, P., Sonenberg, N. Mextli, a novel eIF4E-binding protein from Drosophila. A. Dros. Res. Conf., 2008; 49: 37-42.
- Hiltner, L. Uber neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter bessonderer Berucksichtigung der Gr¨undung und Brache. Arb. Dtsch. Lwirtsch. Ges. Berl., 1904; 98: 59–78.
- Holt, J.G., Krieng, N., Sneath, P., Staley, J., Williams, S.T. Bergey’s Manual of Determinative Bacteriology. (9nd ed.). Baltimore., 1994.
- Janzen, R., Rood, S., Dormaar, J., McGill, W. Azospirillum brasilense produces gibberellin in pure culture on chemical-defined medium in co-culture on straw. Soil Biol. Biochem., 1992; 24: 1061-1064.
- Jha, B., Pragash, M., Cletus, J., Raman, G., Sakthivel, N. Simultaneous phosphate solubilization potential antifungal activity of new fluorescent pseudomonad strains, Pseudomonas aeruginosa, P. plecoglossicida P. Mosselii. World J. Microbiol. Biotecnol., 2009; 25: 573–581.
- Jiang, F., Chen, L., Belimov, A., Shaposhnikov, A., Gong, F., Meng, X., Hartung, W., Jeschke, D., Davies, W., Dodd, I. Multiple impacts of the plant growth-promoting rhizobacterium Variovorax paradoxus 5C-2 on nutrient ABA relations of Pisum sativum. J. Exp. Bot., 2012; 63: 6421–6430.
- Kloepper, J., Leong, J., Teintze,M., Schroth, M. Enhancing plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature., 1980a; 286: 885–886.
- Kloepper, J., Leong, J., Teintze, M., Schroth, M. Pseudomonassiderophores: A mechanism explaining disease-suppressive soils. Curr Microbiol., 1980b; 4: 317–320.
- Kloepper, J., Schroth, M. Plant growthpromoting rhizobacteria on radishes. In: Proceeding of the 4th International Conference on Plant Pathogenic Bacteria. Vol. II Station de Pathologie Vegetale et Phytobacteriologie, INRA, Angers, France, 1978. pp 879-882
- Kumar, D., Prasad, R., Kishore, K., Rao, E. Effect of Azolla (Azolla pinnata) based concentrate mixture on nutrient utilization in buffalo bulls. Indian J. Anim. Res., 2012; 46: 268-271.
- Li, Z., Chang, C., Lin, L., Li, Y., An, Q. A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett. Appl. Microbiol., 2011; 53: 178-185.
- Lugtenberg, B., Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol., 2009; 63: 541–56.
- Pandey, S., Ghosh, P., Ghosh, S., De, T., Maiti T. Role of Heavy metal resistant Ochrobactrum spp. And Bacillus Spp. Strains in bioremediation of a rice cultivar and their PGPR like activities. J. Microbiol., 2013; 51: 11-17.
- Piccoli, P., Lucangeli, C., Schneider, G., Bottini, R. Hydrolysis of [17,17- 2H2] gibberellin A20–glucoside [17,17- 2H2] gibberellin A20–glucosyl ester by Azospirillum lipoferum cultured in a nitrogen-free biotin based chemically-defined medium. Plant Growth Reg. 1997; 23: 179-182.
- Rani, R., Sivakumar, K.. Physico-chemical parameters and phytoplankton richness In certain ponds of Chidambaram, Cuddalore district of Tami Nadu. Int J. Res. Environ. Sci. Technol. 2012; 2: 35-44
- Sharma, H., Sharma, K., Crouch, J. Genetic transformation of crops for insect resistance: Potential and limitations. Cri Reviews in Plant Sci., 2004; 23: 47–72.
- Sharma, P., Khanna, V., Kumari, P. Efficacy of aminocyclopropane-1-carboxylic acid (ACC)-deaminase-producing rhizobacteria in ameliorating water stress in chickpea under axenic conditions. Afr. J. Microbiol. Res., 2013; 7: 5749-5757.
- Singh N, Pandey, P., Dubey, R., Maheshwari, D. Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii by rhizosphere competent Bacillus subtilis BN1. World J. Microbiol. Biotechnol.., 2008; 24: 1669–1679.
- Somasegaran, P., Hoben, H. Handbook of Rhizobia. Methods in Legume-Rhizobium Technology. Springer-Verlag, New York, NY. 1994. pp 193.
- Srinivas, T., Sridevi, M., Mallaiah, K. Effect of pesticides on rhizobium and nodulation of green gram Vigna ratida (L.) Wilczek. ICFAI J. Life Sci., 2008; 2: 36-44.
- Tien, T.M., Gaskins, M., Hubbell, D. Plant growth substances produced by zospirillum brasilenseand Their Effect on the Growth of Pearl Millet (Pennisetum americanum L.) Appl. Environ. Microbiol., 1979; l37: 1016-1037.
- Tilak, K., Ranganayaki, N., Pal, K., De, R., Saxena, A., Shekhar, C., Mittal, S., Tripathi, A., Johri, B. Diversity of plant growth soil health-supporting bacteria. Curr. Sci., 2005; 89: 136–150.
- Voisard, C., Keel, C., Haas, D., Defago, G. Cyanide production by Pseudomonas fluorescens helps suppress black roots rot of tobacco under gnotobiotic conditions. EMBO J., 1989; 8: 351-358. .
- Woese, C.R. Bacterial Evolution. Microbiol. rev., 1987; 51: 221-271.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


