ISSN: 0973-7510
E-ISSN: 2581-690X
The aim of the current study was to evaluate the antibacterial activity of Chlorella vulgaris extracts against Stenotrophomonas maltophilia CSK1 isolated from a diseased cucumber fruit. To this end, water, methanol and ethanol were used separately as solvents to extract the active compounds from the dried Chlorella vulgaris. S. maltophilia CSK1 was identified using the 16S rRNA gene sequencing at 99.93% of identity level. The identity of the bacterial strain CSK1 was confirmed by phylogenetic analyses. CSK1 was clearly clustered with the reference strain, S. maltophilia MTCC 434 (JALV01000036). The antibiosis of the extract against S. maltophilia CSK1 was estimated using agar well diffusion method. Unlike water and ethanolic extracts, the methanolic one exhibited an inhibition of CSK1 (zone of inhibition 13.4 mm), indicating the antimicrobial activity of this extract. No zone of inhibition was formed around ethanolic and water extracts, indicating that both exhibited no-observed effect against the growth of CSK1. This can be attributed to the active constituents in the extract which may varies based on various factors including the polarity of solvent used in the extraction process. The inhibitory effect of the methanolic extract against the S. maltophilia CSK1 obtained from the diseased cucumber fruit could open the door for potential application of such active ingredient within this extract against this bacteria in future.
Chlorella vulgaris, Stenotrophomonas maltophilia, antimicrobial activity
Microalgae are a very diverse group of photosynthetic microbes that inhabit various ecological niches such as fresh and marine water bodies, agricultural soils, deserts and sediments. Microalgae represent a valuable resource for useful metabolites that have a wide range of applications in different fields including industry, medicine and agriculture. It has been reported that microalgae produce a vast amount of various bioactive compounds such as antimicrobial agents, enzymes, immunostimulants.1,2 Chemically, the majority of these substances were verified to be belonged to peptides, fatty acids, alkaloids, and polyketides.
Chlorella vulagris Beijerinck is a common eukaryotic unicellular microalga that has many useful applications. It also has a wide array of biological and pharmacological properties important for human health.3 For example, due to its high protein content, C. vulgaris could be used as a valuable food source in many Asian courtiers particularly in Japan. In addition, this species could produce valuable bioactive compounds. Biological resources particularly microalgae received a considerable attention of researchers worldwide as a potential source for novel antimicrobial substances.2 Chlorellin was the first antimicrobial compound produced by Chlorella that inhibited Gram positive and negative bacteria. Parsiguine was another novel antimicrobial substance the antimicrobial activity that was produced by microalgae.2
Cucumber (Cucumis sativus L.) is an important plant worldwide; its fruit is edible or cooked in many countries. Taxonomically, it belongs to Cucurbitaceae. Cucumber fruits have many important health benefits. It has been reported that cucumber fruits exhibited a significant in vivo antiaging effects. This is likely attributed to the increased content of vitamin C, glutathione peroxidase activity and total phenolic compounds.4 Cucumber fruits also played a protective role against alloxan-induced Diabetes Mellitus.5
Despite these benefits of cucumber fruits, shriveling, water loss and susceptibility to phytopathogens limit the cucumber quantity after harvesting. Many cucumber diseases are reported to be attributed to various microbial agents, e.g., Pseudomonas syringae6, Xanthomonas campestris and Erwinia tracheiphila,7 Additionally, potentially pathogenic bacteria have been reported in cucumber fruits in Riyadh region. The authors explained this by the irrigation with sewage water, the air during harvest and transportation as the expected means for contamination of cucumber with microbes.8
Stenotrophomonas maltophilia is an obligate aerobic, Gram-negative rod, which form endospores. S. maltophilia is an ubiquitous bacterium that inhabit various ecological niches. It has been isolated from vegetables,9 drinking water supply10 (Gomes et al7) pesticide-contaminated soils and Cucumber rhizosphere.11 Furthermore, S. maltophilia is plant-associated and has been isolated from tomatoes (Solanum lycopersicum), potatoes (S. tuberosum), cucumber (Cucumis sativus), various weeds, and other plants.12 S. maltophilia underwent a number of changes in the genus name.13 As per the World Health Organization, S. maltophilia is viewed as a potent multidrug, resistant, less-virulent organism in particular, in hospital settings.14 This bacterium is the causal agent of potato brown rot.15 It also exhibited the ability to cause maceration of Lettuce (Lactuca sativa) and Red mustard greens (Brassica juncea) vegetables9 due to hydrolytic enzymes.
Studies that focus on the microalgae in Al-Ahsa region are very little compared with the ubiquity, diversity and potential applications of this group of microorganisms. It has recently been reported that new natural antimicrobial compounds are existed in the methanolic extract of C. sorokiniana UKM2, Chlorella. sp. and showed a potential activities against many pathogenic bacterial strains such as Pseudomonas aeruginosa, P. aeruginosa strains, Staphylococcus epidermidis, S. aureus, Escherichia coli, Methicillin-resistant S. aureus, Shigella sonnei, Bacillus subtilis, B. subtilis and Serratia marcescens16. Nonetheless, little is known about the C. vulagris in this region and its potential application for production of bioactive compounds. Thus, the objective of the current work was to assess the antimicrobial potential of the microalga, Chlorella vulagris, against many bacterial pathogens.
Source and mass production of Chlorella vulgaris
C. vulgaris was obtained from College of agricultural and food sciences, King Faisal University, Saudi Arabia. In order to obtain an appropriate mass for the subsequent assays, C. vulgaris was inoculated in 500 ml-flasks (10 flasks) containing 250 ml of the BG-11 medium. Constant illumination was used at 60 μE m -2 sec -1 intensity with white fluorescent lamps. The flasks were incubated in illuminated cabinet at 28 °C, for 10 days. After incubation, the cultures were harvested by centrifugation at 10,000 rpm for 15 min.17
Preparation of C. vulgaris Extracts
Exactly, 1 gram of air-dried C. vulgaris was ground separately in a ceramic pestle and mortar with 10 ml of methanol and ethanol, for 10 min in presence of about 10 ml liquid nitrogen. Repeating addition of liquid nitrogen and thawing was carried out three times, to break the cell walls. After grinding, the culture supernatant and solvent extracts were dried at 40 °C for 48h, and then stored at in -20 °C, until use.
Testing the antimicrobial activities of the algal extract
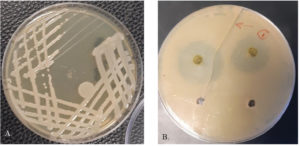
The antimicrobial activities of the methanolic, ethanolic and water extracts of the C. vulgaris was tested using agar-diffusion assay. One ml of each test bacterial strain CSK1 (~ 106 CFU.ml-1) was seeded into Muller Hinton agar. Using sterilized core borer, wells of 6 mm in diameter were cut aseptically, and 50 uL of the relevant extract was added. The plates were carefully placed in incubator at 28℃ for 24h. After incubation, the plates were visually checked for appearance of zone of inhibition around the wells as a positive result (Fig. 1B). The diameter of the formed zone was measured using a ruler. The experiment was done in triplicate.
Determination of the Minimum Inhibitory Concentration (MIC)
The MIC of the algal extract was determined using the extract solution 500 mg/ ml as the stock solution. Two-fold serial dilution from the stock solution was made. Fifty microliters from each dilution was carefully placed in the seed Muller Hinton agar plates. Appropriate control using sterile water and ethanol solution without the extracts was used. The plates were incubated as mentioned previously, at 28℃ for 24 hrs. The lowest concentration, which did not show any growth of tested bacteria by macroscopic evaluation, was determined as the MIC. MIC represents the lowest concentration that inhibits the organism’s growth.
Isolation of the bacterial strain (CSK1) from infected Cucumber
Samples of rotten cucumber fruits were collected from commercial local markets in Al-Ahsa. Bacterial isolation was carried out by direct scratching the infected parts of the Cucumber fruit using sterilized inoculation needle and streaking onto on King’s B medium (20g peptone (BioBasic- Canada), 1.5g K2HPO4 (Merck- Germany). The inoculated plates were incubated at 28°C for 48h (Fig. 1A). Single discrete colonies were picked out and re-streaked onto fresh on King’s B medium plates and preserved for and cryopreservation in 20% glycerol. protocol.
Fig. 1. Growth S. maltophilia CSK1 (A) and antibacterial activity of methanolic extract of C. vulgaris against S. maltophilia CSK1 (B).
Characterization of strain CSK1 using API 20E kit
CSK1 was characterized biochemically using an API20E strip (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s protocol. API20E strip consists of 20 different chemical substrates to be utilized by the tested bacterial strain. Inoculated strips were incubated at 30 °C and results were scored after 24 h.
Identification of the bacterial strain CSK1
Extraction of genomic DNA from the bacterial strain CSK1
In order to extract genomic DNA, bacterial suspension from was mid-exponential phase culture of CSK1 was boiled in InstaGene Matrix (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. The extracted DNA (1-μL aliquot) was used as template DNA for the subsequent PCR amplification of the 16S rRNA gene.
16S rRNA gene amplification using the PCR
In order to amplify the 16S rRNA gene of CSK1, a total of 20 μL PCR reaction in presence of the universal primers; 27F and 1492R18 was used as described earlier.19 Negative control where no-added template and positive control where genomic DNA from E. coli was used as an appropriate negative and positive controls, respectively. The PCR products were checked on agarose gel and the size was estimated using 1 K DNA ladder. DNA will be visualized on a UV trans–illuminator with the Gel Documentation System. The PCR amplicons with right size was proceeded for subsequent steps.
16S rRNA sequencing
Big Dye terminator cycle sequencing kit (Applied BioSystems, Foster City, CA) was used determine the sequences of the 16S rRNA gene of the bacterial isolate. Products of the sequencing were resolved on an Applied Biosystems model 3730XL automated DNA sequencing system.
Phylogenetic analysis
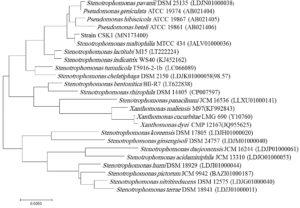
Comparative 16SrRNA gene sequencing analysis of the bacterial strain was carried out using BLAST search against the GenBank nr/nt database, NCBI. Closely related 16S rRNA sequences of recognized species exhibited high level of similarity to the bacterial strain CSK1 were retrieved from NCBI. Phylogenetic relationships were obtained using the Maximum Likelihood method, using the freely available MEGA5.0220. The 16SrRNA sequence of the strain CSK1 was placed in the deposited in the NCBI database under specific accession number (MN173400).
Statistical analysis
Data was analyzed using one-way analysis of variance (ANOVA). P value < 0.05 was considered as significant. SPSS 15.0 was employed for statistical analysis.
Microalgae have become one of the valuable, renewable and ecofriendly sources for novel antibacterial agents due to their short generation time. In this study, three different extracts of the green microalga, C. vulgaris using methanol, ethanol and water as solvents, were tested for its ability to inhibit growth S. maltophilia CSK1. CSK1 was isolated from rotten Cucumber fruit (Cucumis sativu). CSK1 was identified as S. maltophilia using 16S rRNA gene sequencing.
Table (1):
A list of closely related bacterial strain to the CSK1 based on the similarity percentage.
No |
Name |
Strain |
Accession number |
Similarity (%) |
|---|---|---|---|---|
1 |
S. maltophilia |
MTCC 434 |
JALV01000036 |
99.93 |
2 |
S. pavanii |
DSM 25135 |
LDJN01000038 |
99.45 |
3 |
Pseudomonas hibiscicola |
ATCC 19867 |
AB021405 |
99.34 |
4 |
P. beteli |
ATCC 19861 |
AB021406 |
99.29 |
5 |
P. s geniculata |
ATCC 19374 |
AB021404 |
99.29 |
6 |
S. lactitubi |
M15 |
LT222224 |
99.18 |
7 |
S. indicatrix |
WS40 |
KJ452162 |
98.83 |
8 |
S. tumulicola |
T5916-2-1b |
LC066089 |
98.63 |
9 |
S. chelatiphaga |
DSM 21508 |
LDJK01000058 |
98.56 |
10 |
S. bentonitica |
BII-R7 |
LT622838 |
98.19 |
11 |
S. rhizophila |
DSM 14405 |
CP007597 |
97.82 |
API 20E stands for Analytical Profile Index is a biochemical panel developed by the bioMerieux, Inc. for the identification and differentiation of bacterial strains. API 20E strip holds twenty mini-test chambers containing dehydrated media having chemically defined compositions for each test. They usually detect enzymatic activity by the inoculated organisms. CSK1 utilized 55 % (11 out of 20) of the different biochemical testers existed in the API20E strip. CSK1 had the ability to consume all the carbohydrate tested except the rhamnose and inositol, a polyol carbohydrate, (Table 3). Furthermore, CSK1 consumed urea, Na pyruvate and charcoal gelatin. Nonetheless, CSK1 was unable to utilize ortho-nitrophenyl-β-galactoside (ONPG), Arginine, Lysine and Ornithine. These findings in are in general agreement with those recorded in Stenotrophomonas spp. 15 (Messiha et al13) and highlight that CSK1 has versatile enzymatic systems that enable it to utilize various testers but that it devoid some others.
Table (2):
Antimicrobial activity of the C. vulgaris extracts against.
| No | Zone of inhibition (mm) | ||
|---|---|---|---|
| Methanol | Ethanol | Water | |
| S. maltophilia CSK1 | 13 ± 0.4 | 0 | 0 |
| MIC | 60 ± 5 | ND | ND |
ND: not determined
Sequencing the 16s rRNA gene that codes for the small subunit of the ribosome found in bacteria is a perfect target to complete your taxonomy or phylogeny studies18 (Weisburg et al.23) The 16S rRNA sequences of the strain CSK1 was obtained and deposited in the NCBI database under the following accession number (MN173400). BLAST analysis of the 16S rRNA sequences of the CSK1 and other closely related bacterial species showed that CSK1is clearly clustered with the reference strain, S. maltophilia MTCC 434 (JALV01000036). CSK1displayed a substantially high homology (99.93%) homology of the 16S rRNA gene sequences with S. maltophilia MTCC 434 (Accession no. JALV01000036), (Table 1 and Fig. 2). Inferring phylogenetic relationships and evolutionary history among bacterial and archaeal taxa relies on the 16S rRNA genes, in addition to other housekeeping genes. CSK1 was clustered within S. maltophilia clade as inferred by the maximum likelihood method. Phylogenetic trees generated with the neighbor-joining method using the MEGA7 software were similar to the trees constructed with the maximum likelihood method, thus confirming the robust positioning of the bacterial isolate. Therefore, it can be concluded that the 16S rDNA sequence-based phylogenetic tree analysis revealed that CSK1 belonged to the Stenotrophomonas sp. and grouped with S. maltophilia MTCC 434 (Fig. 2), providing a robustness of the identification of the strain. An efficient tool, which is commonly used in identifying and classifying of prokaryotic microorganisms, is 16S rRNA gene sequencing. This is due to its high variability in sequence and widespread in nature among various bacterial and archaeal taxa.
CSK1 was isolated from rotten cucumber fruits and identified as S. maltophilia. Comparable results were introduced by Messiha et al.15 who declared that S. maltophilia was causal agent of potato brown rot. Furthermore, the results in the present study are consistent with those recorded by Lee et al. 9 who noticed that S. maltophilia was responsible for maceration of Lettuce (Lactuca sativa) and Red mustard greens (Brassica juncea) vegetables. Such ability to cause maceration is attributed to the extracellular hydrolyases secreted by the S. maltophilia. Chlorella extract was able to inhibit the bacterial growth of S. maltophilia According to the World Health Organization, S. maltophilia is viewed as a potent multidrug, resistant, less-virulent organism in particular, in hospital settings.14
Methanol extraction is a well-documented method to obtain active antimicrobial agents from microalgae.21 As can be seen in Table (2), the diameter of the zone of inhibition around the S. maltophilia CSK1 indicating the antimicrobial activity of this extract. This could be attributed to the presence of biologically active compounds in this extract that via unknown mechanism ceased the growth of CSK1. In an earlier study, phytochemical screening of C. vulgaris revealed that various bioactive compounds such as flavonoids, terpenoids and tannin were existed in the ethanol extract.
Table (3):
Results of API20 of the strain CSK1.
Number |
Substrate |
Enzyme involved/reaction tested |
Result |
|---|---|---|---|
1 |
ONPG * |
Beta-galactosidase |
– |
2 |
Arginine (ADH) |
Arginine dihydrolase |
− |
3 |
Lysine (LDC) |
Lysine decarboxylase |
− |
4 |
Ornithine (ODC) |
Ornithine decarboxylase |
− |
5 |
Citrate (CIT) |
Citrate utilization |
− |
6 |
Na thiosulfate (H2S) |
H2S production |
− |
7 |
Urea (URE) |
Urea hydrolysis |
+ |
8 |
Tryptophan (TDA) |
Deaminase |
− |
9 |
Tryptophan (IND) |
Indole production |
+ |
10 |
Na pyruvate (VP) |
Acetoin production |
+ |
11 |
Charcoal gelatin (GEL) |
Gelatinase |
+ |
12 |
Glucose (GLU) |
Fermentation/oxidation |
+ |
13 |
Mannitol (MAN) |
Fermentation/oxidation |
+ |
14 |
Inositol (INO) |
Fermentation/oxidation |
− |
15 |
Sorbitol (SOR) |
Fermentation/oxidation |
+ |
16 |
Rhamnose (RHA) |
Fermentation/oxidation |
− |
17 |
Sucrose (SAC) |
Fermentation/oxidation |
+ |
18 |
Melibiose (MEL) |
Fermentation/oxidation |
+ |
19 |
Amygdalin (AMY) |
Fermentation/oxidation |
+ |
20 |
Arabinose (ARA) |
Fermentation/oxidation |
+ |
a+ positive result, CSK1 was able to utilize the substrate; − the negative result, CSK1 was unable to utilize the substrate ONPG Ortho-nitrophenyl-β-galactoside
The minimal inhibitory concentration of the methanolic extract of C. vulgaris is 60 µg ml. It is generally accepted, that MIC represents the lowest concentration that inhibits the organism’s growth. Consequently, the lower the MIC value the more effective the antimicrobial activity of the extract is. Determination of MIC is of pivotal importance since insufficient level of a certain extract or a drug to inhibit bacterial growth, can lead to evolution of potent resistant bacterial strains against this drug or extract. Our results are slightly higher than those recorded by Ahamed et al. 22 who denoted that MIC obtained from Oscillatoria acuminata NTAPC05 against S. maltophilia B929 was (50 μg ml− 1). The MIC value varies from one microalga species to another and on the type of the test organisms.
No zone of inhibition was formed around ethanolic and water extracts, indicating that both exhibited no-observed effect against the growth of the S. maltophilia CSK1. These findings coincide with that obtained by Velichkova et al. 23 who found that methanolic and ethanolic extracts exhibited potent and diverse antibacterial and antifungal activities. In that study, ethanolic extract showed antibacterial activity against E. coli and Pseudomonas aeruginosa whereas methanolic extract of the same microalga species did not show antibiosis. Our results can be explained by the fact that the active constituent in an extract depends on many factors including the polarity of solvent used in the extraction process. For example, water-loving (hydrophilic) compounds are extracted in polar solvent such as ethanol and methanol. Bacterial susceptibility to the active constituents in C. vulagris varies from one species to another. It is well-known that Gram-negative bacteria have an additional outer membrane that is composed of phospholipids and glycolipid lipopolysaccharides, in their cell wall structure. Such layer acts as a protecting-wall against toxic agents.
It can be concluded that the methanolic extract of C. vulagris exhibited a potential inhibitory effect against the S. maltophilia CSK1 obtained from the diseased cucumber fruit. This finding opens the door for promising bicontrol applications of extracts from C. vulagris in future.
ACKNOWLEDGMENTS
The authors would like to thank the Deanship of Scientific Research at King Faisal University, KSA for the financial support. The authors are also thankful to the Biological Sciences Department, King Faisal University for the facilities offered to carry out this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AK put the conception and design, conducted the experimental work, discussed the results and wrote the manuscript. MA helped in collecting samples, experimental work and revising the manuscript. YA cultivated the microalga for mass production. All authors read and approved the manuscript for publication.
FUNDING
This work was supported by Deanship of Scientific Research at King Faisal University, KSA with Grant No. 150195.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The datasets generated and/or analyzed during the current study are available in the NCBI database repository, Accession No (MN173400) of the sequences of the 16S rRNA gene for the strain CSK1.
- Ahamed AAP, Rasheed MU, Noorani KPM, et al. In vitro antibacterial activity of MGDG-palmitoyl from Oscillatoria acuminata NTAPC05 against extended-spectrum β-lactamase producers. J Antibiot. 2017;70(6):754-762.
Crossref - Al-Anazi KA, Al-Jasser AM. Infections caused by Stenotrophomonas maltophilia in recipients of hematopoietic stem cell transplantation. Front Oncol. 2014;4:232.
Crossref - Barghbani R, Rezaei K, Javanshir A. Investigating the effects of several parameters on the growth of Chlorella vulgaris using Taguchi’s experimental approach. Int J Biotechnol Wellness Ind. 2012;1(2):128-133.
- Brooke JS. New strategies against Stenotrophomonas maltophilia: a serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert. Rev Anti Infect Ther. 2014;12(1):1-4.
Crossref - Cui H, Yang G, Jiang J, Zhang P, Gu W. Biological effects of PAS TiO2sol on disease control and photosynthesisin cucumber (‘Cucumis sativus L.’). Aust J Crop Sci. 2013;7(1):99-103.
- Dixit Y, Kar A. Protective Role of Three Vegetable Peels in Alloxan Induced Diabetes Mellitus in Male Mice. Plant Foods Hum Nutr. 2010;65(3):284-289.
Crossref - Gomes IB, Simoes LC, Simoes M. The effects of emerging environmental contaminants on Stenotrophomonas maltophilia isolated from drinking water in planktonic and sessile states. Sci Total Environ. 2018;643:1348-1356.
Crossref - Islam S, Akanda AM, Prova A, Islam MT, Hossain MM. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front Microbiol. 2016;6:1360.
Crossref - Ji L, GaoW, Wei J, Pu L, Yang J, Guo C. In vivo antioxidant properties of lotus root and cucumber: A pilot comparative study in aged subjects. J Nutr Health Aging. 2015;19(7):765-770.
Crossref - Khalifa A, Lee, CG, Ogiso T, et al. Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int J Syst Evol Microbiol. 2015;65(10):3527-3534.
Crossref - Matos J, Cardoso CL, Fale P, Afonso CM, Bandarra NM. Investigation of nutraceutical potential of the microalgae Chlorella vulgaris and Arthrospira platensis. Int J Food Sci Technol. 2020;55(1):303-312.
Crossref - Lee DH, Kim JB, Kim M, et al. Microbiota on spoiled vegetables and their characterization. J Food Prot. 2013;76(8):1350-1358.
Crossref - Messiha NAS, van Diepeningen AD, Farag NS, Abdallah S, Janse JD, van Bruggen A. Stenotrophomonas maltophilia: a new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur J Plant Pathol. 2007;118(3):211-225.
Crossref - Nematian T, Salehi Z, Shakeri A. Conversion of bio-oil extracted from Chlorella vulgaris micro algae to biodiesel via modified superparamagnetic nano-biocatalyst. Renewable Energy. 2020;146(7):1796-1804.
Crossref - Olczak-Woltman H, Bartoszewski G, Madry W, Niemirowicz-Szczytt K. Inheritance of resistance to angular leaf spot (Pseudomonas syringae pv. lachrymans) in cucumber and identification of molecular markers linked to resistance. Plant Pathology. 2009;58(1):145-151.
Crossref - Patil L, Kaliwal BB. Microalga Scenedesmus bajacalifornicus BBKLP-07, a new source of bioactive compounds with in vitro pharmacological applications. Bioprocess Biosyst Eng. 2019;42(6):979-994.
Crossref - Shair OHM, Al-Ssum RM, Hatamleh A. Bacterial contamination in cucumber (cucumis sativus): Evidence From Saudi Arabia. J Pure Appl Microbiol. 2014;8(Spl.edn.2):743-747.
- Shaima AF, Yasin NHM, Ibrahim N, Takriff MS, Gunasekaran D, Ismaeel MYY. Unveiling antimicrobial activity of microalgae Chlorella sorokiniana (UKM2), Chlorella sp.(UKM8) and Scenedesmus sp.(UKM9). Saudi J Biol Sci. 2021.
Crossref - Singh SK, Kaur R, Bansal A, Kapur S, Sundaram S. Biotechnological exploitation of cyanobacteria and microalgae for bioactive compounds. Biotechnological Production of Bioactive Compounds. 2020:221-259.
Crossref - Stoyanova MI, Ganeva DG, Petrov NM, Bogatzevska NS. Stenotrophomonas maltophilia-an Emerging Pathogen of Local Varieties of Tomatoes in Bulgaria. Acta Microbiologica Bulgarica. 2018;34(3):180-186.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5, Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28(10):2731-2739.
Crossref - Velichkova K, Sirakov I, Denev S. In vitro antibacterial effect of Lemna minuta, Chlorella vulgaris and Spirulina sp. Extracts against fish pathogen Aeromonas hydrophila. Aquaculture, Aquarium, Conservation & Legislation. 2019;12(3):936-940.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697-703.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.