ISSN: 0973-7510
E-ISSN: 2581-690X
b-Glucosidase is a class of hydrolytic enzymes that catalyzes the removal of the non-reducing b-D-glucosyl unit from various disaccharides and substituted b-D-glucosides. b-Glucosidase belongs to Glycoside Hydrolase (GH) families 1 and 3 and potentially has many biotechnological applications with thermostable enzymes are preferred over mesophilic homologs in different applications. In the present work, a comparative analysis of physicochemical properties and amino acids composition of 60 (20 mesophilic, 20 thermophilic and 20 hyperthermophilic) b-glucosidases were performed. Multiple sequence alignment and phylogenetic tree analysis were constructed. Analysis of Variance (ANOVA) showed that several physicochemical properties including molecular weight, isoelectric point, number of positively charged amino acids, and extinction coefficient are statistically different among b-glucosidases groups (P<0.05). The analysis also showed that content of amino acids Asp, Gln, Cys, His, and Thr is significantly higher in mesophilic enzymes whereas that of Glu, Lys, Tyr, and Trp is higher in thermo- and hyperthermostable homologs (P<0.05). Overall, nonpolar amino acids were the most abundant amino acids group in b-glucosidase with no significant difference among meso-, thermo-, and hyperthermophilic enzymes. Conversely, the content of polar amino acids is statistically higher (P<0.05) in mesophilic enzymes whereas that of charged and aromatic amino acids is significantly higher (P<0.05) in thermo- and hyperthermophilic counterparts. Finally, multiple regression analysis showed that both polar and aromatic amino acids contribute significantly (P<0.05) to the thermostability. Optimal temperature variation of 53% could be explained by these two groups of amino acids. In conclusion, several amino acids appear to contribute to the thermostability of b-glucosidases and the findings from this study should pave the road toward a better understanding of thermostability of b-glucosidases and protein engineering.
b-Glucosidase, thermostability, amino acid composition, physicochemical properties, ANOVA, Regression.
b-Glucosidase (BGL) is a heterogeneous group of hydrolytic enzymes that catalyzes the removal of the non-reducing b-D-glucosyl terminal unit from a variety of disaccharides, alkyl-b-D-glucosides, aryl-b-D-glucosides and short oligosaccharides1,2. BGLs have found tremendous applications in various biotechnological industries mainly biofuel production, aroma and flavor enhancement, nutritional isoflavone hydrolysis, cassava detoxification, paper deinking, and synthesis of various oligosaccharides and substituted-b-D-glycosides2,3. Hence BGLs have attracted the interest of researchers of this field in the last decade. Additionally, many applications of BGL such as biofuel production require enzymes with exceptional properties such as increased catalytic efficiency, high thermostability, and glucose tolerance4,5. BGLs such as GH 3 BGLs from fungi are sensitive to glucose. However, several reported GH 1 BGLs exhibit excellent glucose tolerance6-10. Thermostability of BGL from GH 1 family is low and the search for thermostable enzymes with glucose tolerance is an important goal of ongoing researches. In this context, on one hand, thermostable enzymes can be obtained through isolation of novel microbes capable of producing thermostable enzymes; which is tedious, time-consuming and cost-intensive approach11. On the other hand, the application of protein engineering principles to design and synthesize thermostable proteins from their mesophilic homologs is the approach of choice toward the development of industrially convenient catalysts3,12.
Elucidation of factors contributing to protein thermostability is the first crucial step for successful protein engineering and catalysts designing for the conversion of mesophilic enzymes to thermophilic counterparts. Several workers of the field have identified some attributes contributing to protein thermostability including hydrophobicity and compactness13-15, shortening of loops16-18, decreased occurrence of thermolabile residues such as Gln, Cys, and Ser13, high content of aromatic amino acids19, high helical content20, increased polar surface area21, hydrogen bonding and electrostatic interactions13,20, high frequency of proline occurrence22, and high disulfide bonds13,23,24. These factors can be determined experimentally or through the analysis of protein sequences and structures using robust computational biology and bioinformatics tools; known as in silico approach. This approach is more attractive because it is cost effective and enables comparison and analysis of large datasets of protein. Detailed comparative analysis of physicochemical properties and amino acids composition of mesophilic, thermophilic, and hyperthermophilic BGLs is lacking. The present study aimed to compare the physicochemical properties and amino acids composition of mesophilic, thermophilic and hyperthermophilic BGLs from GH 1 in an attempt to identify attributes associated with enhanced thermostability of BGLs which may pave the way toward future engineering of BGL.
Data Collection
Different literature databases (e.g., PubMed, ScienceDirect, Springer, Google Scholar) were searched for publications regarding GH1 BGLs. Publications were downloaded and screened for information regarding source organisms, life domain, optimal temperature of enzyme activity and Genbank or UniProt ID (if reported). Only GH1 BGLs which have been characterized for substrate specificity and optimal temperature were selected for further analysis. Protein sequences were retrieved from UniProt (https://www.uniprot.org/) in FASTA format for analysis. Based on reported optimal temperature of enzyme activity, these enzymes were classified into three groups: 1) mesophilic with an optimal temperature between 25-45°C (M-BGLs), 2) thermophilic with an optimal temperature between 50-75°C (T-BGLs), and 3) hyperthermophilic with an optimal activity above 75°C (HT-BGLs).
Deduction of Physicochemical Properties and Amino Acid Compositions
Protein sequences were predicted for the presence of signal peptide using Signal P 4.1 server (http://www.cbs.dtu.dk/services/SignalP/)25, and localization using LocTree 3 and PSORTb (https://rostlab.org/services/loctree3/ and http://www.psort.org/psortb/)26,27. Various physicochemical properties and amino acids composition of protein sequences were also predicted using the EXPASY tool ProtParam (https://web.expasy.org/protparam/)28,29. The physicochemical properties predicted include numbers of amino acids, molecular weight (MW), Isoelectric Points (PI), number of negatively (Asp and Glu) and positively (Lys and Arg) charged residues, extinction coefficient, Instability Index (II), Aliphatic Index (AI) and Grand Average of Hydropathicity (GRAVY).
Sequence Alignment and Phylogenetic Tree Construction
The retrieved sequence of M-BGLs, T-BGLs, and HT-BGLs were aligned using muscle tool for multiple sequence alignment (MSA) at EMBL-EBI (https://www.ebi.ac.uk/Tools/msa/muscle/)30. The alignment was retrieved in FASTA format and edited by BoxShade server (https://embnet.vital-it.ch/software/BOX_form.html)31. Further, MSA was submitted to Phylogeny online tool (http://www.phylogeny.fr/) to construct a phylogenetic tree of selected BGLs sequences32 using the defualt setting (maximum likelihood method, WAG substitution model, bootstrap 16).
Statistical Analysis and Significance Inference
Graphpad Prism 5 was used for calculating statistical parameters of physicochemical properties and amino acids compositions for M-BGLs, T-BGLs, and HT-BGLs. First, analysis of variance (ANOVA) was carried out to find whether there is a significant difference in the means of the parameters of M-BGLs, T-BGLs, and HT-BGLs. The null hypothesis states that there is no significant difference in the means of physicochemical properties and amino acid composition between M-BGLs, T-BGLs, and HT-BGLs. The confidence interval for significance was 95% and P-value <0.05 was considered significant. Next, where ANOVA detected a significant difference, post hoc Tukey’s test was used for multiple comparisons of the means of two groups. Finally, attributes showed a significant difference between M-BGLs, T-BGLs, and HT-BGLs and correlated with the optimal temperature of BGLs activity were used for multiple linear regression analysis to construct a model for optimal temperature prediction from amino acids compositions.
Multiple Sequence Alignment and Phylogenetic Tree Construction
Total sixty GH1 BGL sequences for which experimental optimal temperature has been determined (20 M-BGLs, 20 T-BGLs, and 20 HT-BGLs; Tables 1-3, respectively) were retrieved from the UniProt database. Multiple Sequence Alignment (MSA) analysis revealed that several amino acids motifs are conserved among all GH1 BGLs. b-Glucosidase is a single polypeptide protein that folds to form a GH1 classical (b/a)8 TIM barrel structure comprised of eight a-helices and eight b-strands linked by short loops. GH1 BGL utilizes two key glutamic acid residues as a general acid/base catalyst and nucleophile87. MSA showed the conservation of both Glu residues in all BGLs regardless of optimal temperature. The first Glu residue is the general acid/base conserved at position 166 (for BGL from Humicola insolens (HiBGL) as reference, see supplementary data S. Fig. 1) at conserved motif TXNEP (Thr-X-Asn-Glu-Pro) and the second Glu residue is the nucleophile conserved at position 377 in consensus sequence TENG (Thr-Glu-Asn-Gly)88. The active site is located at C-terminal of the barrel and is made up of two subsites namely glycon binding site (subsite -1) and aglycon binding site (subsite +1). The catalytic acid/base is located at the C-terminal of b-strand 4 and the nucleophile at the C-terminal of the b-strand 788. In HiBGL, glycon binding site (subsite -1) lies at the bottom of the barrel with Gln17, His120, Trp121, Asn165, Tyr308, Trp427, Glu434, Trp435 and Phe443 residues65. MSA showed that these residues are conserved throughout BGL evolutionary history and the side chains of which interact with glycon moiety through both hydrogen and hydrophobic bonds. Conversely, aglycon binding site (subsite +1) is less conserved and is determined by Thr177, Tyr179, Phe325, Leu326, Thr331, Phe333 and Phe348 (in HiBGL numbering) which function as gatekeepers and explain the aglycone broad substrate specificity exhibited by this enzyme89. Moreover, Trp 168 and Leu173 were found to be responsible for glucose tolerance90 and MSA showed that these two residues are conserved among high glucose-tolerant BGLs. The aglycon appeared to anchor by hydrophobic contacts and water-mediated polar bonds91. In contrarily, there are few studies on amino acids/motifs associated with thermostability of BGLs. Tamaki et al. 2014 employed Statistical Coupling Analysis (SCA) to identify several amino acids related to the thermostability of BGL from Spodoptera frugiperda (Sfbgly) (corresponding to Arg27, Pro39, Trp121, Pro167, His211, Pro266, Pro286, Trp435 and Phe443 in HiBGL numbering)92. MSA demonstrated that these residues are conserved and the majority of which are proline or positively charged amino acids. Additionally, these residues appeared to be distributed in the loop segments of BGL whereas, amino acids related to enzyme activity are mainly concentrated around a-helices and b-strands92. Altogether these residues represent a hotspot for BGL engineering in future. However, further studies to identify more amino acids variants and motifs related to the thermostability in M-, T-, and HT-BGL may be required.
Table (1):
Mesophilic GH1 β-Glucosidase with UniProt ID and reported optimal temperature.
Enzyme ID |
UniProt ID |
Source organism |
Domain |
Optima T |
Reference |
|---|---|---|---|---|---|
M-BGL01 |
K0A8J9 |
Exiguobacterium antarcticum B7 |
Bacteria |
30 |
[33] |
M-BGL02 |
O93785 |
Hypocrea jecorina |
Fungi |
40 |
[34] |
M-BGL03 |
A1D6G3 |
Neosartorya fischeri NRRL181 |
Fungi |
40 |
[35] |
M-BGL04 |
F1JZ12 |
Sphingomonas sp. strain 2F2 |
Bacteria |
37 |
[36] |
M-BGL05 |
B9V8P5 |
Micrococcus antarcticus |
Bacteria |
25 |
[37] |
M-BGL06 |
A0A1S5SJM8 |
Unculturable bacterium |
Bacteria |
40 |
[38] |
M-BGL07 |
D5KX75 |
Unculturable bacterium |
Bacteria |
40 |
[8] |
M-BGL08 |
I6YQJ8 |
Unculturable bacterium |
Bacteria |
40 |
[39] |
M-BGL09 |
E6TUY6 |
Bacillus cellulosilyticus |
Bacteria |
40 |
[40] |
M-BGL10 |
I6TNE2 |
Weissella cibaria |
Bacteria |
45 |
[41] |
M-BGL11 |
Q9F3B7 |
Streptomyces coelicolor A3 |
Bacteria |
35 |
[42] |
M-BGL12 |
Q9K440 |
Streptomyces coelicolor A3 |
Bacteria |
35 |
[42] |
M-BGL13 |
D0VLH9 |
Exiguobacterium oxidotolerans |
Bacteria |
35 |
[43] |
M-BGL14 |
J9XU85 |
Bifidobacterium lactis |
Bacteria |
38 |
[44] |
M-BGL15 |
B8HAF9 |
Arthrobacter chlorophenolicus |
Bacteria |
37 |
[45] |
M-BGL16 |
A0A1L3HS62 |
Uncultured bacterium |
Bacteria |
37 |
[46] |
M-BGL17 |
A0A2I2LGB3 |
Uncultured bacterium |
Bacteria |
40 |
[47] |
M-BGL18 |
A0A1W6I0S4 |
Uncultured bacteriuma |
Bacteria |
38 |
[48] |
M-BGL19 |
A6W3B1 |
Marinomonas MWYL1 |
Bacteria |
40 |
[49] |
M-BGL20 |
M4I6Y9 |
Lactococcus sp. FSJ4 |
Bacteria |
40 |
[50] |
a1-18 amino acids were predicted as signal sequence and removed
Table (2):
Thermophilic GH 1 β-Glucosidase with UniProt ID and reported optimal temperature.
Enzyme ID |
UniProt ID |
Source |
Domain |
Optima T |
Reference |
|---|---|---|---|---|---|
T-BGL01 |
HV538882.1 |
Uncultured bacterium |
Bacteria |
75 |
[51] |
T-BGL02 |
A0A0B5ARU7 |
Jeotgalibacillus malaysiensis |
Bacteria |
65 |
[52] |
T-BGL03 |
Q47RE2 |
Thermobifida fusca |
Bacteria |
60 |
[53] |
T-BGL04 |
M5QUM2 |
Anoxybacillus sp. DT3-1 |
Bacteria |
70 |
[10] |
T-BGL05 |
K4I4U1 |
Uncultured bacterium |
Bacteria |
50 |
[54] |
T-BGL06 |
D9TR57 |
Thermoanaerobacterium thermosaccharolyticum |
Bacteria |
70 |
[55] |
T-BGL07 |
A0LR48 |
Acidothermus cellulolyticus |
Bacteria |
70 |
[56] |
T-BGL08 |
A0A220YLM5 |
Alicyclobacillus sp. |
Bacteria |
55 |
[6] |
T-BGL09 |
H0HC94 |
Agrobacterium tumefaciens 5A |
Bacteria |
52 |
[57] |
T-BGL10 |
A0A0H4NXH8 |
Thermoanaerobacterium aotearoense |
Bacteria |
60 |
[9] |
T-BGL11 |
W0LHR5 |
Uncultured bacterium |
Bacteria |
60 |
[58] |
T-BGL12 |
Q65D37 |
Bacillus licheniformis |
Bacteria |
50 |
[59] |
T-BGL13 |
Q608B9 |
Methylococcus capsulatus |
Bacteria |
70 |
[60] |
T-BGL14 |
A4XIG7 |
Caldicellulosiruptor saccharolyticus |
Bacteria |
70 |
[61] |
T-BGL15 |
A0A220IP58 |
Cellulosimicrobium cellulans |
Bacteria |
55 |
[62] |
T-BGL16 |
Q60026 |
Thermoanaerobacter brockii |
Bacteria |
75 |
[63] |
T-BGL17 |
B8CYA8 |
Halothermothrix orenii |
Bacteria |
70 |
[64] |
T-BGL18 |
I3QIG4 |
Bacillus subtilis |
Bacteria |
60 |
[7] |
T-BGL19 |
A0A076JRL8 |
Humicola insolens RP86 |
Fungi |
60 |
[65] |
T-BGL20 |
H8XVY6 |
Paecilomyces thermophila |
Fungi |
55 |
[66] |
Table (3):
Hyperthermophilic GH1 β-Glucosidase with UniProt ID and reported optimal temperature.
Enzyme ID |
UniProt ID |
Source organism |
Domain |
Optima T |
Reference |
|---|---|---|---|---|---|
HT-BGL01 |
E7FHY4 |
Pyrococcus furiosus |
Archaea |
100 |
[67] |
HT-BGL02 |
O08324 |
Thermococcus sp. |
Archaea |
78 |
[68] |
HT-BGL03 |
Q08638 |
Thermotoga maritima |
Bacteria |
95 |
[69] |
HT-BGL04 |
F7YX70 |
Thermotoga thermarum |
Bacteria |
90 |
[70] |
HT-BGL05 |
G8YZD7 |
Fervidobacterium islandicum |
Bacteria |
90 |
[71] |
HT-BGL06 |
A5IL97 |
Thermotoga petrophila |
Bacteria |
80 |
[72] |
HT-BGL07 |
W8W3B8 |
Uncultured bacterium |
Achaea |
90 |
[73] |
HT-BGL08 |
A0A0A6ZH67 |
Uncultured bacterium |
Bacteria |
90 |
[74] |
HT-BGL09 |
Q746L1 |
Thermus thermophiles HB27 |
Bacteria |
88 |
[75] |
HT-BGL10 |
P22498 |
Sulfolobus solfataricus |
Archaea |
90 |
[76] |
HT-BGL11 |
B9K7M5 |
Thermotoga neapolitana |
Bacteria |
95 |
[77] |
HT-BGL12 |
B8E1X9 |
Dictyoglomus turgidum |
Bacteria |
80 |
[78] |
HT-BGL13 |
D3Y2V4 |
Thermoanaerobacter ethanolicus |
Bacteria |
80 |
[79] |
HT-BGL14 |
A8WAC9 |
Thermus thermophiles HJ6 |
Bacteria |
90 |
[80] |
HT-BGL15 |
P10482 |
Caldocellum saccharolyticum |
Bacteria |
85 |
[81] |
HT-BGL16 |
Q9YGA8 |
Thermosphaera aggregans |
Archaea |
85 |
[82] |
HT-BGL17 |
D9PZ08 |
Acidilobus saccharovorans |
Archaea |
93 |
[83] |
HT-BGL18 |
Q9YGB8 |
Pyrococcus kodakaraensis |
Archaea |
100 |
[84] |
HT-BGL19 |
P14288 |
Sulfolobus acidocaldarius |
Archaea |
85 |
[85] |
HT-BGL20 |
A0A0A7RBQ4 |
Thermococcus pacificus P-4 |
Archaea |
75 |
[86] |
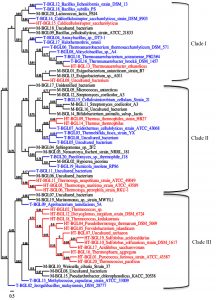
Fig. 1. Phylogenetic tree of mesophilic (Black, M-BGL), thermophilic (blue, T-BGL) and hyperthermophilic (red, HT-BGL) b-glucosidases from bacteria, archaea, and fungi. This phylogenetic tree was constructed using Phylogeny tool.
Multiple sequence alignment was used to construct a phylogenetic tree to visualize the evolutionary relationship between M-, T-, and HT-BGLs. BGLs were clustered into three major clades (Fig. 1). Clade I was dominated by T-BGLs (9 sequences, 52.9%) followed by M-BGLs (5, 29.4%) and HT-BGLs (3, 17.7%). Clade II was dominated by M-BGLs (10, 43.5%) followed by T-BGLs (8, 34.8%) and HT-BGLs (5, 21.7%). Both mesophilic and thermophilic fungal BGLs analyzed were clustered together in this clade suggesting their bacterial origin. Clade III was dominated by HT-BGLs from both bacteria and archaea (12, 60%) followed by M-BGLs (5, 25%) and thermostable BGLs (3, 15%). Clustering of mesophilic, thermophilic and hyperthermophilic BGLs together indicates the existence of structural and functional similarities. The clustering of mesophilic and thermophilic protein is in agreement with previous reports93,94. Similarly, HT-BGLs from both archaea and bacteria were also clustered together in clade III indicating the structural similarity among them. Archaeal and bacterial proteins have been clustered together in several phylogenetic tree analyses95,96. This is because many proteins distinguishing these two domains belong to information processing proteins such as DNA replicating enzymes, and transcription and translation associated protein97.
Table (4):
Statistical analysis of physicochemical properties of GH1 β-glucosidases.
| Physicochemical properties | Average | ANOVA Statistics | Tukey’s multiple comparison test, significant? | ||||
|---|---|---|---|---|---|---|---|
| M-BG | T-BG | HT-BG | F Value P Value | M vs T | M vs HT | T vs HTa | |
| No. of amino acid residues | 455.1±21.71 | 462.35±15.27 | 461.35±25.81 | 0.676 0.51 | No | No | No |
| Molecular Weight (Da) | 51390.9±2223.73 | 52518.51±1365.76 | 53321.49±3079.78 | 3.463 0.038 | No | Yes | No |
| Theoretical PI | 5.28±0.81 | 5.27±0.31 | 5.80±0.42 | 5.882 0.005 | No | Yes | Yes |
| No. of Negatively charged residue (Asp+Glu) | 62.05±8.65 | 66.55±5.58 | 63.75±6.18 | 2.15 0.126 | No | No | No |
| No of Positively charged residue (Arg+Lys) | 41.8±5.47 | 47.45±7.28 | 53.75±4.87 | 20.11 0.000 | Yes | Yes | Yes |
| Extinction Coefficients | 104913±14154.08 | 111164.75±10943.43 | 124925.75±14061.23 | 12.146 0.000 | No | Yes | Yes |
| Instability Index
II |
32.76±5.25 | 32.11±4.20 | 32.57±5.97 | 0.084 0.92 | No | No | No |
| Aliphatic Index
AI |
77.71±6.74 | 77.75±4.44 | 79.23±4.66 | 0.577 0.565 | No | No | Yes |
| Grand average of hydropathicity (GRAVY) | -0.37±0.13 | -0.4152±0.11 | -0.41675±0.1 | 1.171 0.318 | No | No | No |
a M for M-BGL, T for T-BGL, and HT for HT-BGL
Comparative Analysis of Physicochemical Properties
All GH1 BGLs appear to lack of signal peptide and to localize in the cytoplasm except M-BGL-18 which was predicted to have 18 residues and to localize in the periplasm. GH 1 BGLs are known to be localized in the cytoplasm3. Statistical analysis (ANOVA and followed by Tukey test) demonstrated that MW of HT-BGLs is significantly higher than M-BGLs or T-BGL (P<0.05, Table 4). Increase in the MW of HT-BGLs could be attributed to the higher content of larger amino acids such as Lys, Tyr and Trp and lower content of smaller amino acids such as Gly, Gln, and Cys in HT-BGLs98,99. Similarly, PI is significantly higher in HT-BGLs than M-BGLs and T-BGLs (P<0.05, Table 4). A similar finding was reported for thermostable nitrilase over their mesophilic counterparts95. PI indicates the pH at which the protein has an equal number of positive and negative charges. However, a study on a set of 310 proteins failed to correlate pH or temperature stability with PI100,101. Additionally, the analysis showed that numbers of positively charged amino acids (Lys and Arg) are higher in HT-BGLs than M-BGLs and T-BGLs (P< 0.05, Table 4). Increased content of positively charged amino acids in thermostable BGLs can be postulated to involve in salt bridge formations and thus enhancing protein thermostability102-105. Indeed, there is experimental evidence showing that the redesigning of salt bridge significantly enhanced BGL thermostability106. Finally, the extinction coefficient is also statistically higher in HT-BGLs than M-BGLs and T-BGLs (P<0.05, Table 4). Extinction coefficient reflects aromatic amino acids content (Phe, Tyr, and Trp) which in turn appears to enhance protein thermostability through increasing protein hydrophobicity and packing106. Conversely, number of negatively charged amino acids, AI, II, and GRAVY did not show any statistical difference in their means among M-, T-, and HT-BGLs. Similar findings have been reported for nitrilase/cyanide hydratase family from mesophilic, thermophilic, and hyperthermophilic bacteria95 and serine protease from mesophilic and thermophilic microorganisms107. AI indicates the relative volume occupied by the side chain of hydrophobic amino acids (Ala, Val, Leu, and Ile) and may suggest thermostability of protein. AI was higher for all BGLs analyzed in the present study suggesting their overall stability108.
Table (5):
Statistical analysis of amino acids composition (%) of GH1 β-glucosidases.
| Amino Acid | Average±SD | ANOVA analysis | Tukey multiple comparison, significant? | |||||
|---|---|---|---|---|---|---|---|---|
| M-BGL | T-BGL | HT-BGL | F Value | P value | M vs T | M vs HT | T vs HTa | |
| Ala (A) | 9.02±2.5 | 8.56±2.7 | 7.28±2.2 | 2.605 | 0.083 | No | No | No |
| Arg (R) | 5.47±1.7 | 5.54±1.5 | 5.81±1.8 | 0.231 | 0.795 | No | No | No |
| Asn (N) | 4.23±1.1 | 3.98±1.1 | 4.70±1.4 | 1.792 | 0.176 | No | No | No |
| Asp (D) | 7.58±1.6 | 7.72±1.1 | 6.17±0.9 | 9.690 | 0.000 | No | Yes | Yes |
| Cys (C) | 0.95±0.7 | 0.66±0.4 | 0.40±0.4 | 5.463 | 0.007 | No | Yes | No |
| Gln (Q) | 3.18±1.1 | 2.38±1.0 | 1.86±0.7 | 10.439 | 0.000 | Yes | Yes | No |
| Glu (E) | 6.06±1.5 | 6.68±1.5 | 7.67±1.1 | 7.245 | 0.001 | No | Yes | No |
| Gly (G) | 8.2±1.3 | 8.47±0.8 | 7.65±0.9 | 3.54 | 0.035 | No | No | Yes |
| His (H) | 3.21±0.8 | 3.19±0.9 | 2.63±0.5 | 4.04 | 0.03 | No | Yes | Yes |
| Ile (I) | 5.2±1.5 | 5.39±1.8 | 5.8±1.6 | 0.723 | 0.490 | No | No | No |
| Leu (L) | 8.09±1.4 | 8.04±1.1 | 7.75±1.5 | 0.401 | 0.671 | No | No | No |
| Lys (K) | 3.71±1.8 | 4.76±2.7 | 5.84±2.1 | 4.54 | 0.015 | No | Yes | No |
| Met (M) | 1.81±0.7 | 1.89±0.7 | 2.1±0.8 | 0.859 | 0.429 | No | No | No |
| Phe (F) | 4.67±1.4 | 4.54±1.1 | 5.02±0.8 | 0.982 | 0.381 | No | No | No |
| Pro (P) | 4.89±1.1 | 4.89±1.2 | 5.21±1.2 | 0.537 | 0.586 | No | No | No |
| Ser (S) | 4.89±1.1 | 4.65±1.0 | 4.71±1.3 | 0.236 | 0.79 | No | No | No |
| Thr (T) | 5.29±0.8 | 4.48±1.1 | 3.48±0.7 | 19.996 | 0.000 | Yes | Yes | Yes |
| Trp (W) | 2.84±0.5 | 2.88±0.5 | 3.28±0.3 | 5.68 | 0.006 | No | Yes | Yes |
| Tyr (Y) | 4.93±0.8 | 5.5±0.9 | 6.08±0.5 | 11.554 | 0.000 | No | Yes | Yes |
| Val (V) | 5.84±1.2 | 5.81±1.3 | 6.63±1.2 | 2.9 | 0.063 | No | No | No |
| nonpolar | 43.03±3.7 | 42.57±3.2 | 42.40±3.1 | 0.186 | 0.831 | No | No | No |
| polar | 21.74±2.3 | 19.54±1.6 | 17.78±2.3 | 16.807 | 0.000 | Yes | Yes | Yes |
| charged | 22.81±2.2 | 24.86±2.3 | 25.48±1.8 | 8.058 | 0.001 | Yes | Yes | No |
| aromatic | 12.44±1.9 | 13.00±1.8 | 14.37±0.9 | 7.033 | 0.002 | No | Yes | Yes |
a M for M-BGL, T for T-BGL, and HT for HT-BGL
Comparative Analysis of Amino Acids Composition
ANOVA analysis demonstrated that Asp, Cys, Gln, His and Thr are significantly higher in M-BGLs than HT-BGLs and T-BGL homologs (P< 0.05, Table 5). These amino acids are unstable at higher temperature and undergo either oxidations or deamination at higher temperature explaining why they are less common in thermostable protein compared to mesophilic homologs22,95,104,109-111. Cys specifically plays a dual role by, on one hand, reducing thermostability through increasing internal cavities and oxidation at a higher temperature and, on the other, increasing thermostability through the formation of disulfide bonds which enhance protein rigidity and stability112. Conversely, Glu, Lys, Trp, and Tyr are significantly higher in HT-BGLs than their T-BGLs and M-BGLs counterparts (P<0.05, Table 5). Glu is negatively charged amino acids common in both exposed and buried region of the protein and involved in electrostatic interactions. Farias et al. (2003) found E+K increased and Q+H decreased in thermostable protein suggesting E+K/Q+H ratio can be used as an indicator of thermal stability113. Similarly, Lys is positively charged amino acid which involves in ionic interactions resulting in enhanced thermo-stability and hence it is more abundant in thermophilic and hyperthermophilic proteins114-116. Furthermore, both Trp and Tyr are aromatic amino acids which are more common in thermostable protein than their mesophilic homologs13,117. Aromatic amino acids contribute to protein thermostability through p-p and cation-p interactions12,118. Gly was significantly higher in T-BGLs than HT-BGLs or M-BGLs homologs (P<0.05, Table 5). Gly is small hydrophobic amino acid responsible for creating void or cavity in the interior of protein thus hyperthermostable protein are evolved to have less Gly content to minimize the cavities which may disturb protein upon temperature increase104,117. The analysis also showed that there is no significant difference in the means of nonpolar amino acids Ala, Ile, Leu, Met, Phe, Pro, Val, and polar amino acid Met, Arg, Asn, and Ser between M-BGL, T-BGL and HT-BGL homologs (P>0.05, Table 5). Ala is the best helix forming residue associated with increased thermostability and packing of the protein119,120. Ile was found to be more common in thermostable compared to mesophilic protein100. Phe is a hydrophobic amino acid that tends to bury inside protein thus was higher in hyperthermophilic protein than their meso- and thermophilic homologues121. Previous research reported that a-helices of thermophilic protein are more stable than those of mesophilic homologs perhaps due to the high abundance of amino acids with greater propensity to form a-helices (Ala, Leu, Arg) and low abundance in b-branch sheet forming residues (Val, Ile, Thr). a-helices of thermostable protein can also be stabilized by interactions between side chains of amino acids such as Glu and Arg119,122,123. Pro has pyrrolidine ring which allows it to have least conformational states and low conformational entropy restricting the configuration of preceding amino acids thus it is more common on rigid and turn conformations and hence reported to be higher in thermophilic protein116. Pro has been used to increase protein thermo-stability and can be considered, here, a potential hotspot to enhance thermostability of BGLs124. Similarly, Met, Asn, and Ser are thermo-labile that undergo either oxidation or deamination (Asn) at elevated temperature and are therefore less common in the thermostable protein125,126. Indeed, the substitution of Ser by Ala in thermophilic protein is widely reported100. Arg is a positively charged residue that participates in electrostatic bond formation to enhance protein stability127,128. The present study cannot justify why the residues such as Ala, Phe, Arg, and Pro which generally contribute to thermostability are not statistically higher in thermostable BGLs than mesophilic one. However, it is important to note that this study compared protein sequences solely from one family (GH1 BGLs) whereas previous studies compared protein sequences from several families; it is well-reported that different protein families adopt different strategies to enhance their thermostability12.
Collectively, nonpolar amino acids (Ala, Gly, Ile, Leu, Met, Pro, Val) were the most abundant amino acids in all BGLs accounting for about 42.5% of total amino acids with no statistical difference in their means between M-, T-, and HT-BGLs (P>0.05, Table.5). Nonpolar amino acids are buried in the interior of protein and influence its hydrophobicity which is the major interacting force responsible for the stability of protein core104,117. Chakravarty et al. (2002) reported that nonpolar amino acids are relatively higher in thermophilic protein than their mesophilic protein114. Conversely, polar amino acids (Asn, Gln, Ser, Thr, His, Cys) are significantly higher in M-BGLs than T-BGLs and HT-BGLs (P<0.05, Table 5). Decrease of polar amino acids in thermostable enzymes contributes to thermostability by minimizing cavities, Gln- and Asn- induced deamidation, and Cys, Ser and Thr oxidation at higher temperatures. This finding is in agreement with previous reports13,125,126,129. In contrary, charged amino acids (Glu, Asp, Lys, Arg) are higher in HT-BGLs and T-BGLs than M-BGLs (P< 0.05, Table 5). Increase of charged amino acids in the thermostable protein was previously reported and appears to mediate protein thermostability through the formation of hydrogen and ionic interactions115,126,130. Finally, aromatic amino acids (Phe, Tyr, Trp) are also significantly higher in HT-BGL than M-BGL and T-BGL analogs (P<0.05). This increase in aromatic amino acids enhances thermostability by increasing hydrophobicity of protein through cation-p and p-p interaction131 and compactness/packing of protein and decreasing cavities106.
Multiple Regression Analysis
As previously demonstrated, the mean numbers of positively, polar, and aromatic amino acids are significantly different between M-, T-, and HT-BGLs with both positively and aromatic amino acids are directly correlated with optimal temperature (r= 0.62 and r= 0.65, respectively) and polar amino acids are negatively correlated (r= -0.55). These variables were used to perform multiple linear regression to determine a model for predicting optimal temperature. However, the positively charged amino acids were excluded from the model because it failed to be a significant predictor as indicated by individual test (P> 0.05). The model was constructed with polar and aromatic amino acids which significantly predicted optimal temperature with R square value of 0.53 indicating that variance in optimal temperature of 53% could be explained by the variation of these two groups of amino acids. This model also has multiple correlation coefficients R of 0.741 indicating that a high-quality prediction of this model. Additionally, b- coefficient indicates that aromatic amino acids (Trp+Tyr) contributed more to predicting optimal temperature than polar amino acids (Table 6). Of note, the low prediction value of this model (53%) is because thermostability cannot be solely predicted from the primary sequences of protein132.
Table (6):
Multiple regression analysis of polar and aromatic amino acids for Optimal temperature prediction.
Variable |
Coefficient |
Std. Error |
β |
T value |
Sig. |
|---|---|---|---|---|---|
Intercept |
31.018 |
25.984 |
1.194 |
0.238 |
|
Polar Amino Acids |
-3.008 |
0.765 |
-0.374 |
-3.934 |
0.000 |
Tyr + Trp |
10.687 |
1.942 |
0.523 |
5.504 |
0.000 |
Thermostable BGLs differ from their mesophilic counterparts in several physicochemical properties such as molecular weight, isoelectric points, positively charged amino acids, and extinction coefficient. The high abundance of nonpolar amino acids in all BGLs may indicate general stability of BGLs. Additionally, increase in aromatic amino acids (Tyr and Trp) and decrease in polar amino acids (Gln, His, Thr, Cys) contributes significantly to BGL thermostability probably by combined mechanisms of increased hydrophobicity and decreases cavities of globular proteins. Charged amino acids (Lys and Glu) may also contribute to BGL thermostability through the formation of ionic bonds. Overall, these amino acids may be targeted through protein engineering for the conversion of mesophilic BGLs to their thermostable analogs. However, thermostability cannot be predicted solely from amino acids composition since the spatial arrangement of amino acids and structural feature of protein influence protein thermostability. Therefore, future analysis should focus on characterizing amino acids motifs and secondary structure of mesophilic and thermophilic BGLs to elucidate more attributes associated with thermostability. Furthermore, benefiting from a large number of X-ray crystallographic structures of BGLs elucidated to date, a comparative analysis of 3D structures may provide a deep insight into the difference between mesophilic and thermophilic BGLs thus paving the road toward successful protein engineering of this industrially valuable enzyme.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of beta-glucosidases. 3 Biotech, 2016; 6: 315-328.
Crossref - Ahmed, A.; Aslam, M.; Ashraf, M.; ul-Hassan Nasim, F.; Batool, K.; Bibi, A. Microbial -Glucosidases: Screening, Characterization, Cloning and Applications. Journal of Applied & Environmental Microbiology, 2017; 5: 57-73.
Crossref - Ahmed, A.; Nasim, F. u.-H.; Batool, K.; Bibi, A. Microbial Beta-Glucosidase: Sources, Production and Applications. Journal of Applied & Environmental Microbio. Ogy., 2017; 5: 31-46.
Crossref - Sorensen, A, L beck, M, L beck, PS, Ahring, BK. Fungal Beta-glucosidases: a bottleneck in industrial use of lignocellulosic materials. Biomolecules, 2013; 3: 612-631.
Crossref - Rani, V.; M, S.; Tiwari, R.; Nain, L.; Arora, A. Beta-Glucosidase: Key Enzyme in Determining Efficiency of Cellulase and Biomass Hydrolysis. Journal of Bioprocesses & Biotechniques, 2014; 5: 197-205.
Crossref - Cao, H.; Zhang, Y.; Shi, P.; Ma, R.; Yang, H.; Xia, W. et al. A highly glucose-tolerant GH1 beta-glucosidase with greater conversion rate of soybean isoflavones in monogastric animals. Journal of Indian Microbiology & Biotechnology, 2018; 9: 2018-2040.
Crossref - Chamoli, S.; Kumar, P.; Navani, N.K.; Verma, A.K. Secretory expression, characterization and docking study of glucose-tolerant beta-glucosidase from B. subtilis. International Journal of Biological Macromolecules, 2016; 85: 425-433.
Crossref - Fang, Z.; Fang, W.; Liu, J.; Hong, Y.; Peng, H.; Zhang, X. et al. Cloning and characterization of a beta-glucosidase from marine microbial metagenome with excellent glucose tolerance. Journal of Microbiology & Biotechnology, 2010; 20: 1351-1358.
Crossref - Yang, F.; Yang, X.; Li, Z.; Du, C.; Wang, J.; Li, S. Overexpression and characterization of a glucose-tolerant beta-glucosidase from T. aotearoense with high specific activity for cellobiose. Applied Microbiology & Biotechnology, 2015; 99: 8903-8915.
Crossref - Chan, C.S.; Sin, L.L.; Chan, K.G.; Shamsir, M.S.; Manan, F.A.; Sani, R.K. et al. Characterization of a glucose-tolerant beta-glucosidase from Anoxybacillus sp. DT3-1. Biotechnology Biofuels, 2016; 9: 174.
Crossref - Mohammad, B.T.; Al Daghistani, H.I.; Jaouani, A.; Abdel-Latif, S.; Kennes, C. Isolation and Characterization of Thermophilic Bacteria from Jordanian Hot Springs: Bacillus licheniformis and Thermomonas hydrothermalis Isolates as Potential Producers of Thermostable Enzymes. International Journal of Microbiology, 2017; Article No: 6943952.
Crossref - Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein thermostability engineering. RSC Advances, 2016; 6: 115252-115270.
Crossref - Kumar, S.; Tsai, C.-J.; Nussinov, R. Factors enhancing protein thermostability. Protein Engineering, Design and Selection, 2000; 13: 179-191.
Crossref - Chirakkal, H.; Ford, G.C.; Moir, A. Analysis of a conserved hydrophobic pocket important for the thermostability of Bacillus pumilus chloramphenicol acetyltransferase (CAT-86). Protein Engineering, 2001; 14: 161-166.
Crossref - Janecek, S. Does the increased hydrophobicity of the interior and hydrophilicity of the exterior of an enzyme structure reflect its increased thermostability? International Journal of Biological Macromolecules, 1993; 15: 317-318.
Crossref - Balasco, N.; Esposito, L.; De Simone, A.; Vitagliano, L. Role of loops connecting secondary structure elements in the stabilization of proteins isolated from thermophilic organisms. Protein Science, 2013; 22: 1016-1023.
Crossref - Dagan, S.; Hagai, T.; Gavrilov, Y.; Kapon, R.; Levy, Y.; Reich, Z. Stabilization of a protein conferred by an increase in folded state entropy. Proceeding National Academy of Science (U.S.A), 2013; 110: 10628-10633.
Crossref - Yokota, K.; Satou, K.; Ohki, S.-y. Comparative analysis of protein thermostability: Differences in amino acid content and substitution at the surfaces and in the core regions of thermophilic and mesophilic proteins. Science and Technology of Advanced Materials, 2006; 7: 255-262.
Crossref - Szilagyi, A.; Zavodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure, 2000; 8: 493-504.
Crossref - Coleman, R.G.; Sharp, K.A. Shape and evolution of thermostable protein structure. Proteins, 2010; 78: 420-433.
Crossref - Vetriani, C.; Maeder, D.L.; Tolliday, N.; Yip, K.S.; Stillman, T.J.; Britton, K.L. et al. Protein thermostability above 100 degreesC: a key role for ionic interactions. Proceeding National Academy of Science (U.S.A), 1998; 95: 12300-12305.
Crossref - Panja, A.S.; Bandopadhyay, B.; Maiti, S. Protein Thermostability Is Owing to Their Preferences to Non-Polar Smaller Volume Amino Acids, Variations in Residual Physico-Chemical Properties and More Salt-Bridges. PLoS One, 2015; 10: e0131495.
Crossref - Watanabe, K.; Chishiro, K.; Kitamura, K.; Suzuki, Y. Proline residues responsible for thermostability occur with high frequency in the loop regions of an extremely thermostable oligo-1,6-glucosidase from Bacillus thermo-glucosidasius KP1006. Journal of Biological Chemistry, 1991; 266: 24287-24294.
- Yang, H.M.; Yao, B.; Meng, K.; Wang, Y.R.; Bai, Y.G.; Wu, N.F. Introduction of a disulfide bridge enhances the thermostability of a Streptomyces olivaceoviridis xylanase mutant. Journal of Industrial Microbiology & Biotechnology, 2007; 34: 213-218.
Crossref - Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods, 2011; 8: 785-796.
Crossref - Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R. et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics, 2010; 26: 1608-1615.
Crossref - Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N. et al. LocTree3 prediction of localization. Nucleic Acids Research, 2014; 42: W350-W355.
Crossref - Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S. e.; Wilkins, M. R.; Appel, R. D. et al. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook, Walker, J.M., Ed. Humana Press, Totowa, NJ, 2005; 571-607;
Crossref - Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P. W.; Kayastha, A. M. et al. MFPPI – Multi FASTA ProtParam Interface. Bioinformation, 2016; 12: 74-77.
Crossref - Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 2004; 32: 1792-1797.
Crossref - Gille, C.; Birgit, W.; Gille, A. Sequence alignment visualization in HTML5 without Java. Bioinformatics, 2014; 30: 121-132.
Crossref - Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research, 2008; 36: W465-W469.
Crossref - Crespim, E.; Zanphorlin, L.M.; de Souza, F.H.; Diogo, J.A.; Gazolla, A.C.; Machado, C.B.; et al. A novel cold-adapted and glucose-tolerant GH1 beta-glucosidase from Exiguobacterium antarcticum B7. International Journal Biological Macromolecules, 2016; 82: 375-380.
Crossref - Jeng, W.Y.; Wang, N.C.; Lin, M.H.; Lin, C.T.; Liaw, Y.C.; Chang, W.J.; et al. Structural and functional analysis of three beta-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis. J. Structural Biology, 2011; 173: 46-56.
Crossref - Kalyani, D.; Lee, K.M.; Tiwari, M.K.; Ramachandran, P.; Kim, H.; Kim, I.W. et al. Characterization of a recombinant aryl beta-glucosidase from Neosartorya fischeri NRRL181. Applied Microbiology & Biotechnology, 2012; 94: 413-423.
Crossref - Wang, L.; Liu, Q.M.; Sung, B.H.; An, D.S.; Lee, H.G.; Kim, S.G. et al. Bioconversion of ginsenosides Rb(1), Rb(2), Rc and Rd by novel beta-glucosidase hydrolyzing outer 3-O glycoside from Sphingomonas sp. 2F2: cloning, expression, and enzyme characterization. Journal of Biotechnology, 2011; 156: 125-133.
Crossref - Fan, H.X.; Miao, L.L.; Liu, Y.; Liu, H.C.; Liu, Z.P. Gene cloning and characterization of a cold-adapted beta-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microbial Technology, 2011; 49: 94-99.
Crossref - Suwan, E.; Arthornthurasuk, S.; Kongsaeree Prachumporn, T. A metagenomic approach to discover a novel b-glucosidase from bovine rumens. Pure & Applied Chemistry, 2017; 89: 941-950.
Crossref - Li, G.; Jiang, Y.; Fan, X. J.; Liu, Y. H. Molecular cloning and characterization of a novel beta-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library. Bioresource Technology, 2012; 123: 15-22.
Crossref - Wu, J.; Geng, A.; Xie, R.; Wang, H.; Sun, J. Characterization of cold adapted and ethanol tolerant b-glucosidase from Bacillus cellulosilyticus and its application for directed hydrolysis of cellobiose to ethanol. International Journal of Biological Macromolecules, 2018; 109: 872-879.
Crossref - Lee, K.W.; Han, N.S.; Kim, J.H. Purification and characterization of beta-glucosidase from Weissella cibaria 37. Journal Microbiology & Biotechnology, 2012; 22: 1705-1713.
Crossref - Gu, M.Z.; Wang, J.C.; Liu, W.B.; Zhou, Y.; Ye, B.C. Expression and displaying of beta-glucosidase from Streptomyces coelicolor A3 in Escherichia coli. Applied Biochemistry & Biotechnology, 2013; 170: 1713-1723.
Crossref - Chen, S.; Hong, Y.; Shao, Z.; Liu, Z. A cold-active b-glucosidase (Bgl1C) from a sea bacteria Exiguobacterium oxidotolerans A011. World journal of Microbiology & Biotechnology, 2010; 26: 1427-1435.
Crossref - Youn, S.Y.; Park, M.S.; Ji, G.E. Identification of the beta-glucosidase gene from Bifidobacterium animalis subsp. lactis and its expression in B. bifidum BGN4. Journal of Microbiology & Biotechnology, 2012; 22: 1714-1723.
Crossref - Park, M.K.; Cui, C.H.; Park, S.C.; Park, S.K.; Kim, J.K.; Jung, M.S. et al. Characterization of recombinant beta-glucosidase from Arthrobacter chlorophenolicus and biotransformation of ginsenosides Rb1, Rb 2, Rc, and Rd. Journal of Microbiology, 2014; 52: 399-406.
Crossref - Gomes-Pepe, E.S.; Machado Sierra, E.G.; Pereira, M.R.; Castellane, T.C.; Lemos, E.G. Bg10: A Novel Metagenomics Alcohol-Tolerant and Glucose-Stimulated GH1 ss-Glucosidase Suitable for Lactose-Free Milk Preparation. PLoS One, 2016; 11: e0167932.
Crossref - Toyama, D.; de Morais, M.A.B.; Ramos, F.C.; Zanphorlin, L.M.; Tonoli, C.C.C.; Balula, A.F. et al. A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil). Biochimica Biophysica Acta: Proteins & Proteomics, 2018; 4: 569-579.
Crossref - Zhang, L.; Fu, Q.; Li, W.; Wang, B.; Yin, X.; Liu, S. et al. Identification and characterization of a novel beta-glucosidase via metagenomic analysis of Bursaphelenchus xylophilus and its microbial flora. Scientific Reports, 2017; 7: 14850.
Crossref - Zhao, W.; Peng, R.; Xiong, A.; Fu, X.; Tian, Y.; Yao, Q. Expression and characterization of a cold-active and xylose-stimulated beta-glucosidase from Marinomonas MWYL1 in Escherichia coli. Molecular Biology Report, 2012; 39: 2937-2943.
Crossref - Fang, S.; Chang, J.; Lee, Y.S.; Guo, W.; Choi, Y.L.; Zhou, Y. Cloning and characterization of a new broadspecific beta-glucosidase from Lactococcus sp. FSJ4. World Journal of Microbiology and Biotechnology, 2014; 30: 213-223.
Crossref - Uchiyama, T.; Miyazaki, K.; Yaoi, K. Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity. Journal of Biological Chemisty, 2013; 288: 18325-18334.
Crossref - Liew, K.J.; Lim, L.; Woo, H.Y.; Chan, K.G.; Shamsir, M.S.; Goh, K.M. Purification and characterization of a novel GH1 beta-glucosidase from Jeotgalibacillus malaysiensis. International Journal of Biological Macromolecules, 2018; 115: 1094-1102.
Crossref - Chen, W.-L.; Yang, Y.-M.; Guo, G.-W.; Chen, C.-Y.; Huang, Y.-C.; Liu, W.-H. et al. Over-Expression of the Thermobifida fusca b-Glucosidase in a Yarrowia lipolytica Transformant to Degrade Soybean Isoflavones. Catalysts, 2018; 8: 24.
Crossref - Lu, J.; Du, L.; Wei, Y.; Hu, Y.; Huang, R. Expression and characterization of a novel highly glucose-tolerant beta-glucosidase from a soil metagenome. Acta BioChimica et Biophysica Sinica, 2013; 45: 664-673.
Crossref - Pei, J.; Pang, Q.; Zhao, L.; Fan, S.; Shi, H. Thermoanaerobacterium thermosaccharo-lyticum beta-glucosidase: a glucose-tolerant enzyme with high specific activity for cellobiose. Biotechnology Biofuels, 2012; 5: 31.
Crossref - Li, Y.; Bu, M.; Chen, P.; Li, X.; Chen, C.; Gao, G. et al. Characterization of a Thermophilic Monosaccharide Stimulated b-Glucosidase from Acidothermus cellulolyticus. Chemical Research at Chinese Universities, 2018; 34: 212-220.
Crossref - Goswami, S.; Gupta, N.; Datta, S. Using the beta-glucosidase catalyzed reaction product glucose to improve the ionic liquid tolerance of beta-glucosidases. Biotechnology Biofuels, 2016; 9: 72.
Crossref - Biver,S.; Stroobants, A.; Portetelle, D.; Vandenbol, M. Two promising alkaline beta-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose. Journal of Industrial Microbiology and Biotechnology, 2014; 41: 479-488.
Crossref - Zahoor, S.; Javed, M.M.; Aftab, M.N.; Ikram-ul-Haq. Cloning and expression of b-glucosidase gene from Bacillus licheniformis into E. coli BL 21 (DE3). Biologia, 2011; 66: 213-220.
Crossref - Sathe, S.S.; Soni, S.; Ranvir, V.P.; Choudhari, V.G.; Odaneth, A.A.; Lali, A.M.; Chandrayan, S.K. Heterologous expression and biochemical studies of a thermostable glucose tolerant beta-glucosidase from Methylococcus capsulatus (bath strain). International Journal of Biological Macromolecules, 2017; 102: 805-812.
Crossref - Hong, M.R.; Kim, Y.S.; Park, C.S.; Lee, J.K.; Oh, D.K. Characterization of a recombinant beta-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus. Journal of Bioscience and Bioengineering, 2009; 108: 36-40.
Crossref - Xu, F.; Yao, J.; Hu, Y.; Wang, J.; Zhao, T.; Zhou, Y. et al. Cloning, expression and biochemical characterization of a GH1 b-glucosidase from Cellulosimicrobium cellulans AU – Yuan, Ye. Biocatalalysis and Biotransformation, 2018; 36: 362-371.
Crossref - Breves, R.; Bronnenmeier, K.; Wild, N.; Lottspeich, F.; Staudenbauer, W.L.; Hofemeister, J. Genes encoding two different beta-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Applied and Environmental of Microbiology, 1997; 63: 3902-3910.
- Hassan, N.; Nguyen, T.H.; Intanon, M.; Kori, L.D.; Patel, B.K.; Haltrich, D. et al. Biochemical and structural characterization of a thermostable beta-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis. Applied Microbiology and Biotechnology, 2015; 99: 1731-1744.
Crossref - Souza, F.H.M.; Meleiro, L.P.; Machado, C.B.; Zimbardi, A.L.R.L.; Maldonado, R.F.; Souza, T.A.C. B. et al. Gene cloning, expression and biochemical characterization of a glucose- and xylose-stimulated b-glucosidase from Humicola insolens RP86. Journal of Molecular Catalaysis B: Enzymatics, 2014; 106: 1-10.
Crossref - Yang, S.; Hua, C.; Yan, Q.; Li, Y.; Jiang, Z. Biochemical properties of a novel glycoside hydrolase family 1 beta-glucosidase (PtBglu1) from Paecilomyces thermophila expressed in Pichia pastoris. Carbohydrate Polymers, 2013; 92: 784-791.
Crossref - Voorhorst, W.G.; Eggen, R.I.; Luesink, E.J.; de Vos, W.M. Characterization of the celB gene coding for beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus and its expression and site-directed mutation in Escherichia coli. Journal of Bacteriology, 1995; 177: 7105-7111.
Crossref - Sinha, S.K.; Datta, S. beta-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Applied Microbiology and Biotechnology, 2016; 100: 8399-8409.
Crossref - Mehmood, M.A.; Shahid, I.; Hussain, K.; Latif, F.; Rajoka, M.I. Thermodynamic properties of the beta-glucosidase from Thermotoga maritima extend the upper limit of thermophilicity. Protein Peptide Letters, 2014; 21: 1282-1288.
- Zhao, L.; Xie, J.; Zhang, X.; Cao, F.; Pei, J. Overexpression and characterization of a glucose-tolerant b-glucosidase from Thermotoga thermarum DSM 5069T with high catalytic efficiency of ginsenoside Rb1 to Rd. Journal Molecular Catalysis B: Enzymatics, 2013; 95: 62-69.
Crossref - Jabbour, D.; Klippel, B.; Antranikian, G. A novel thermostable and glucose-tolerant beta-glucosidase from Fervidobacterium islandicum. Applied Microbiology and Biotechnology, 2012; 93: 1947-1956.
Crossref - Cota, J.; Correa, T.L.; Damasio, A.R.; Diogo, J.A.; Hoffmam, Z.B.; Garcia, W.; et al. Comparative analysis of three hyperthermophilic GH1 and GH3 family members with industrial potential. New Biotechnology, 2015; 32: 13-20.
Crossref - Schroder, C.; Elleuche, S.; Blank, S.; Antranikian, G. Characterization of a heat-active archaeal beta-glucosidase from a hydrothermal spring metagenome. Enzyme Microbial Technology, 2014; 57: 48-54.
Crossref - Wang, Q.; Qian, C.; Zhang, X.Z.; Liu, N.; Yan, X.; Zhou, Z. Characterization of a novel thermostable beta-glucosidase from a metagenomic library of termite gut. Enzyme Microbial Technology, 2012; 51: 319-324.
Crossref - Dion, M.; Fourage, L.; Hallet, J.N.; Colas, B. Cloning and expression of a beta-glycosidase gene from Thermus thermophilus. Sequence and biochemical characterization of the encoded enzyme. Glycoconjugates Journal, 1999; 16: 27-37.
Crossref - Kim, B.N.; Yeom, S.J.; Kim, Y.S.; Oh, D.K. Characterization of a beta-glucosidase from Sulfolobus solfataricus for isoflavone glycosides. Biotechnology Letters, 2012; 34: 125-129.
Crossref - Park, T.H.; Choi, K.W.; Park, C.S.; Lee, S.B.; Kang, H. Y.; Shon, K.J. et al. Substrate specificity and transglycosylation catalyzed by a thermostable beta-glucosidase from marine hyperthermophile Thermotoga neapolitana. Applied Microbiology & Biotechnology, 2005; 69: 411-422.
Crossref - Fusco, F.A.; Fiorentino, G.; Pedone, E.; Contursi, P.; Bartolucci, S.; Limauro, D. Biochemical characterization of a novel thermostable beta-glucosidase from Dictyoglomus turgidum. International Journal of Biological Macromolecules, 2018; 113: 783-791.
Crossref - Song, X.; Xue, Y.; Wang, Q.; Wu, X. Comparison of three thermostable beta-glucosidases for application in the hydrolysis of soybean isoflavone glycosides. Journal of Agricultural and Food Chemistry, 2011; 59: 1954-1961.
Crossref - Gu, N.Y.; Kim, J.L.; Kim, H.J.; You, D.J.; Kim, H.W.; Jeon, S.J. Gene cloning and enzymatic properties of hyperthermostable beta-glycosidase from Thermus thermophilus HJ6. Journal Bioscience and Bioengineering, 2009; 107: 21-26.
Crossref - Love, D.R.; Streiff, M.B. Molecular Cloning of a b-Glucosidase Gene from an Extremely Thermophilic Anaerobe in E. coli and B. subtilis. Bio-Technology, 1987; 5: 384-387.
Crossref - Chi, Y.I.; Martinez-Cruz, L.A.; Jancarik, J.; Swanson, R.V.; Robertson, D.E.; Kim, S.H. Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Letters, 1999; 445: 375-383.
Crossref - Gumerov, V.M.; Rakitin, A.L.; Mardanov, A.V.; Ravin, N.V. A Novel Highly Thermostable Multifunctional Beta-Glycosidase from Crenarchaeon Acidilobus saccharovorans. Archaea, 2015; Article NO. 978632.
Crossref - Ezaki, S.; Miyaoku, K.; Nishi, K.; Tanaka, T.; Fujiwara, S.; Takagi, M.; et al. Gene analysis and enzymatic properties of thermostable beta-glycosidase from Pyrococcus kodakaraensis KOD1. Journal of Bioscience and Bioengineering, 1999; 88: 130-135.
Crossref - Noh, K.H.; Oh, D.K. Production of the rare ginsenosides compound K, compound Y, and compound Mc by a thermostable beta-glycosidase from Sulfolobus acidocaldarius. Biological and Pharmaceutical Bulletin, 2009; 32, 1830-1835.
Crossref - Kim, Y.J.; Eun Lee, J.; Lee, H.S.; Kwon, K.K.; Gyun Kang, S.; Lee, J.-H. Novel substrate specificity of a thermostable Beta-glucosidase from the hyperthermophilic archaeon, Thermococcus pacificus P-4. Korean Journal of Microbiology, 2015; 51: 68-74.
Crossref - Berrin, J.-G.; Czjzek, M.; Kroon, P.A.; McLauchlan, W. R.; Puigserver, A.; Williamson, G.; et al. Substrate (aglycone) specificity of human cytosolic beta-glucosidase. The Biochemical Journal, 2003; 373: 41-48.
Crossref - Santos, C.A.; Zanphorlin, L.M.; Crucello, A.; Tonoli, C.C.C.; Ruller, R.; Horta, M.A.C.; et al. Crystal structure and biochemical characterization of the recombinant ThBgl, a GH1 b-glucosidase overexpressed in Trichoderma harzianum under biomass degradation conditions. Biotechnology Biofuel, 2016; 9: 71.
Crossref - Michalska, K.; Tan, K.; Li, H.; Hatzos-Skintges, C.; Bearden, J.; Babnigg, G.; et al. GH1-family 6-P-b-glucosidases from human microbiome lactic acid bacteria. Acta Crystallographic Section D: Biological Crystallography, 2013; 69: 451-463.
Crossref - De Giuseppe, P.O.; Souza Tde, A.; Souza, F.H.; Zanphorlin, L.M.; Machado, C.B.; Ward, R.J.; et al. Structural basis for glucose tolerance in GH1 beta-glucosidases. Acta Crystallographic Section D: Biological Crystallography, 2014; 70: 1631-1639.
Crossref - Zanphorlin, L.M.; de Giuseppe, P.O.; Honorato, R.V.; Tonoli, C.C.; Fattori, J.; Crespim, E.; et al. Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7. Scientific Reports, 2016; 6: 23776.
Crossref - Tamaki, F.K.; Textor, L.C.; Polikarpov, I.; Marana, S. R. Sets of covariant residues modulate the activity and thermal stability of GH1 beta-glucosidases. PLoS One, 2014; 9: e96627.
Crossref - Xia, W.; Bai, Y.; Cui, Y.; Xu, X.; Qian, L.; Shi, P.; et al. Functional diversity of family 3 beta-glucosidases from thermophilic cellulolytic fungus Humicola insolens Y1. Scientific Reports, 2016; 6: 27062.
Crossref - Sood, S.; Sharma, N. Microbial Carboxylesterases: An Insight into Thermal Adaptation Using In Silico Approach. Journal of Proteomics and Bioinformatics, 2016; 9: 131-136.
Crossref - Devi, S.; Sharma, N.; Savitri; Bhalla, T.C. Comparative analysis of amino acid sequences from mesophiles and thermophiles in respective of carbon-nitrogen hydrolase family. 3 Biotech, 2013; 3: 491-507.
Crossref - Yan, S.; Wu, G. Analysis on evolutionary relationship of amylases from archaea, bacteria and eukaryota. World Journal of Microbiology and Biotechnology, 2016; 32: 24.
Crossref - Sarmiento, F.; Long, F.; Cann, I.; Whitman, W.B. Diversity of the DNA replication system in the Archaea domain. Archaea, 2014; Article NO. 675946.
Crossref - Fields, P.A. Review: Protein function at thermal extremes: balancing stability and flexibility. Comparative Biochemistry and Physiology – Part A: Molecular & Integrative Physiology, 2001; 129: 417-431.
Crossref - Sadeghi, M.; Naderi-Manesh, H.; Zarrabi, M. Ranjbar, B., Effective factors in thermostability of thermophilic proteins. Biophysical Chemistry, 2006; 119: 256-270.
Crossref - Tompa, D.R.; Gromiha, M.M.; Saraboji, K. Contribution of main chain and side chain atoms and their locations to the stability of thermophilic proteins. Journal of Molecular Graphics Modelling, 2016; 64: 85-93.
Crossref - Talley, K.; Alexov, E. On the pH-optimum of activity and stability of proteins. Proteins, 2010; 78: 2699-2706.
Crossref - Vogt, G.; Woell, S.; Argos, P. Protein thermal stability, hydrogen bonds, and ion pairs. Journal of Molecular Biology, 1997; 269: 631-643.
Crossref - Zhou, X.X.; Wang, Y.B.; Pan, Y.J.; Li, W.F. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids, 2008; 34: 25-33.
Crossref - Elcock, A.H. The stability of salt bridges at high temperatures: implications for hyperthermo-philic proteins. Journal of Molecular Biology, 1998; 284: 489-502.
Crossref - Lee, C.W.; Wang, H.J.; Hwang, J.K.; Tseng, C.P. Protein thermal stability enhancement by designing salt bridges: a combined computational and experimental study. PLoS One, 2014; 9: e112751.
Crossref - Madhusudan Makwana, K.; Mahalakshmi, R. Implications of aromatic-aromatic interactions: From protein structures to peptide models. Protein Science, 2015; 24: 1920-1933.
Crossref - Raj, T.; Sharma, N.; Savitri; Bhalla, T., Bacterial Serine Proteases: Computational and Statistical Approach to Understand Temperature Adaptability. Journal of Proteomics and Bioinformatics, 2016; 10: 329-334.
Crossref - Lu, B.; Wang, G.; Huang, P. A comparison of amino acid composition of proteins from thermophiles and mesophiles. Wei Sheng Wu Xue Bao, 1998; 38: 20-25.
- Sharma, N.; Kushwaha, R.; J.S, S.; Bhalla, T. In Silico Analysis of Amino Acid Sequences in Relation to Specificity and Physiochemical Properties of Some Microbial Nitrilases. Journal of Proteomics and Bioinformatics, 2009; 2: 185-192.
- Kumwenda, B.; Litthauer, D.; Bishop, O.T.; Reva, O. Analysis of protein thermostability enhancing factors in industrially important thermus bacteria species. Evolutionary Bioinformatics Online, 2013; 9: 327-342.
Crossref - Pack, S.P.; Yoo, Y.J. Protein thermostability: structure-based difference of amino acid between thermophilic and mesophilic proteins. Journal of Biotechnology, 2004; 111: 269-277.
Crossref - Tatara, Y.; Yoshida, T.; Ichishima, E.A single free cysteine residue and disulfide bond contribute to the thermostability of Aspergillus saitoi 1,2-alpha-mannosidase. Bioscience, Biotechnology and Biochemistry, 2005; 69: 2101-2108.
Crossref - Farias, S.T.; Bonato, M.C. Preferred amino acids and thermostability. Genetics and Molecular Research, 2003; 2: 383-393.
- Chakravarty, S.; Varadarajan, R. Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry, 2002; 41: 8152-8161.
Crossref - Trivedi, S.; Gehlot, H. S.; Rao, S. R. Protein thermostability in Archaea and Eubacteria. Genetics and Molecular Research, 2006; 5: 816-827.
- Yang, H.; Liu, L.; Li, J.; Chen, J.; Du, G. Rational Design to Improve Protein Thermostability: Recent Advances and Prospects. ChemBioEng Review, 2015; 2: 87-94.
Crossref - Das, R.; Gerstein, M. The stability of thermophilic proteins: a study based on comprehensive genome comparison. Functional Integrated Genomics, 2000; 1: 76-88.
Crossref - Chakrabartty, A.; Kortemme, T.; Baldwin, R.L. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Science, 1994; 3: 843-852.
Crossref - Malleshappa Gowder, S.; Chatterjee, J.; Chaudhuri, T.; Paul, K. Prediction and analysis of surface hydrophobic residues in tertiary structure of proteins. Scientific World Journal, 2014; Articcle NO. 971258.
Crossref - Yakimov, A.P.; Afanaseva, A.S.; Khodorkovskiy, M.A.; Petukhov, M. G. Design of Stable alpha-Helical Peptides and Thermostable Proteins in Biotechnology and Biomedicine. Acta Naturae, 2016; 8: 70-81.
Crossref - Petukhov, M.; Kil, Y.; Kuramitsu, S.; Lanzov, V. Insights into thermal resistance of proteins from the intrinsic stability of their alpha-helices. Proteins, 1997; 29: 309-320. https://doi.org/10.1002/(SICI)1097-0134(199711)29:3%3C309::AID-PROT5%3E3.0.CO;2-5
- Facchiano, A.M.; Colonna, G.; Ragone, R. Helix-stabilizing factors and stabilization of thermophilic proteins: an X-ray based study. Protein Engineering, 1999; 11: 753-760.
Crossref - Pace, C.N.; Scholtz, J.M.A helix propensity scale based on experimental studies of peptides and proteins. Biophysics Journal, 1998; 75: 422-427.
Crossref - Platts, L.; Falconer, R.J. Controlling protein stability: Mechanisms revealed using formulations of arginine, glycine and guanidinium HCl with three globular proteins. International Journal of Pharmaceutics, 2015; 486: 131-135.
Crossref - Bhanuramanand, K.; Ahmad, S.; Rao, N.M. Engineering deamidation-susceptible asparagines leads to improved stability to thermal cycling in a lipase. Protein Science, 2014; 23: 1479-1490.
Crossref - Haney, P.J.; Badger, J.H.; Buldak, G.L.; Reich, C.I.; Woese, C.R.; Olsen, G.J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proceeding National Academy of Science (U.S.A), 1999; 96: 3578-3583.
Crossref - Shah, D.; Shaikh, A.R.; Peng, X.; Rajagopalan, R. Effects of arginine on heat-induced aggregation of concentrated protein solutions. Biotechnology Progress, 2011; 27: 513-520.
Crossref - Chakravarty, S.; Varadarajan, R. Elucidation of determinants of protein stability through genome sequence analysis. FEBS Letters, 2000; 470: 65-69.
Crossref - Law, M.J.; Linde, M.E.; Chambers, E.J.; Oubridge, C.; Katsamba, P.S.; Nilsson, L.; et al. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Research, 2006; 34: 275-285.
Crossref - Dougherty, D.A. Cation-pi interactions involving aromatic amino acids. Journal of Nutriton, 2007; 137: 1504S-1508S.
Crossref - Soulby, A.J.; Heal, J.W.; Barrow, M.P.; Roemer, R.A.; O’Connor, P.B. Does deamidation cause protein unfolding? A top-down tandem mass spectrometry study. Protein Science, 2015; 24: 850-860.
Crossref - Miotto, M.; Olimpieri, P.P.; Di Rienzo, L.; Ambrosetti, F.; Corsi, P.; Lepore, R. et al. Insights on protein thermal stability: a graph representation of molecular interactions. Bioinformatics, 2018; 10: 1-9.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.