ISSN: 0973-7510
E-ISSN: 2581-690X
Globally, cervical cancer is the fourth most common cancer among women. After being cloned from a recurring cervical lesion in 1987, Human papillomavirus (HPV) type-45 was identified as a high-risk HPV type. It is the third most common cancer-causing HPV subtype, after HPV-16 and HPV-18. Immunogenic epitopes and structural features provide the most useful information for vaccine development. Computational algorithms provide quick, simple, trustworthy, and cost-efficient methods for predicting immunogenic epitopes. In this study, both B and T cell epitopes have been identified as potential immunogens that can elicit a response from the host system. Three potential B-cell epitopes, i.e., SIAGQYRGQCNTCCDQ, LQEIVLHLEPQNELDP, and DSTVYLPPPSVARVVS, were identified in this study. A potential epitope for E6 (ATLERTEVY) was predicted to 8 MHC-I alleles (HLA-A*30:02, HLA-B*15:01, HLA-A*01:01, HLA-A*26:01, HLA-A*32:01, HLA-B*35:01, HLA-B*58:01, HLA-A*11:01) and for L1 epitope (NVFPIFLQM) was predicted for 4 MHC-I alleles (HLA-A*30:02, HLA-A*32:01, HLA-B*53:01, HLA-B*51:01). To conclude, the epitopes identified here might potentially be useful for developing a cervical cancer vaccine against HPV-45 strains, but in vitro and in vivo trials are needed to validate their safety and efficacy.
Human Papillomavirus, Cervical Cancer, Immunogenic Epitopes, Vaccine, In silico
Globally, cervical cancer ranks as the fourth most frequently occurring malignancy among women population.1 Over 500,000 women worldwide are diagnosed with cervical cancer each year, with low-income nations bearing the burden of mortality.2 Nearly all cervical malignancies contain oncogenic human papillomavirus (HPV) DNA. With the highest universally attributable percentage ever reported for a particular etiology of a major human malignancy, researchers concluded that HPV is an essential element in the development of cervical cancer.3 A working committee of the International Agency for Research on Cancer (IARC) Monographs categorized 14 types of HPV as “carcinogenic to humans” out of 200 different types. While the majority of HPV infections are asymptomatic and are eventually removed by our immune system, the virus can remain in some situations, thus leading to cancer.4
A persistent cervical lesion seen in a woman in the United States led to the discovery of the high-risk (HR) HPV type HPV-45 in 1987. HPV-45 is more frequent in adenocarcinoma of the cervix. After HPV-16 and HPV-18, HPV-45 has been ranked as the third most oncogenic type, which accounts for around 10% of cervical cancer cases.4 The cellular structure of this virus is made up of 8,000 bp of circular double-stranded DNA that contains early regions (E1, E2, E4, E5, E6, E7, and E8) encoding early viral proteins, late regions (L1 and L2) that codes for the capsid proteins, and a non-coding region known as the long control region (LCR), which plays a key role in replication and transcription.3,5
The oncoproteins of genes E6 and E7 are identified as the key causes of HPV-associated cervical cancer; elevated expression of E6 and E7 is necessary for the onset and maintenance of the malignant phenotype.6 p53 and pRb (retinoblastoma) tumour suppressors are inactivated when E6 and E7 genes are expressed, respectively.7 These oncoproteins are tumour-specific antigens, and hence there is no risk of autoimmunity. They are expressed in all the phases of cervical cancer, making them ideal targets for prophylactic vaccination.8 The icosahedral capsid structure is formed by the major capsid protein L1. Charged residues (K and R) are concentrated near the C-terminus, and there is often >60% L1 amino acid sequence homology between HPV variants that infect the genital epithelia. It indicates that the majority of the L1 protein is conserved among different types of HPV.9,10 The protein can self-assemble into an icosahedral capsid by forming 72 pentameric capsomers. Because of its icosahedral form, L1 protein is equally distributed on the surface of the capsid, making it highly immunogenic.11 This protein is capable of forming virus-like particles (VLPs) by self-assembling spontaneously. VLPs that have been assembled are thought to be potent immunogens that B-cells can recognise quickly.12
The comprehensive cervical cancer control strategy involves HPV vaccination as primary prevention, screening and treating precancerous lesions as secondary prevention. The Food and Drug Administration (FDA) has approved the use of three forms of prophylactic vaccines: Cervarix® (bivalent), Gardasil® (quadrivalent), and Gardasil®9 (nonavalent). These vaccines are efficient in protecting against HPV infection and neoplasms. However, they are prophylactic vaccines that offer no therapeutic benefit and have limited benefits in eradicating pre-existing infections. As a result, therapeutic vaccinations are gaining popularity due to their capacity to trigger cell-mediated immune responses and destroy infected cells rather than neutralising antibodies (nAbs).13 All of the aforementioned studies suggest that E6, E7, and L1 are key proteins that can be used as a potential vaccine candidate against HPV-45. Using several bioinformatics tools and programmes, we attempted to examine the E6, E7, and L1 proteins of HPV-45 as a potential vaccine candidate in this work.
Amino acid sequence
E6, E7 and L1 amino acid sequences of HPV-45 having GenBank accession numbers CAA52573.1, CAA52574.1 and CAA52578.1 (Genome ID: X74479), respectively, were retrieved from the NCBI databank.
Sequence analysis
Protparam (http://web.expasy.org/protparam/) is an online web tool used for the analysis of the different physical and chemical characteristics of E6, E7 and L1 protein sequences of HPV-45, including molecular mass, amino acid composition, and atomic composition.14
Prediction of secondary structure
PSIPRED is an internet server (http://bioinf.cs.ucl.ac.uk/psipred) that incorporates two feed-forward neural networks that analyse PSI-BLAST (Position-Specific Iterated-BLAST) output to predict concise and precise secondary structure of E6, E7 and L1 proteins of HPV-45.15
T-cell epitope prediction
Immune Epitope Database (IEDB) is a resource (http://www.iedb.org/) funded by the National Institute of Allergy and Infectious Diseases (NIAID), a division of the National Institutes of Health. This tool was used for the prediction of T-cell epitopes for E6, E7 and L1 protein sequences. The IEDB recommended 2020.09 (NetMHCpan El 4.1) prediction method was used to predict the epitopes for MHC-I alleles, while the IEDB recommended 2.22 prediction method was used for MHC-II alleles. The reference set of HLA alleles were selected for predicting both MHC-I and MHC-II binding in various human populations.16 The antigenicity of the predicted epitopes for MHC-I alleles were calculated using Vaxigen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html).17 IFNepitope web server (http://crdd.osdd.net/raghava/ifnepitope/) was used to investigate the ability of epitopes for MHC-II alleles to stimulate interferon-gamma (IFN-g) production. The parameters for this study were set as IFN-g versus non-IFN-g model and Motif and SVM hybrid algorithms.18
B-cell epitope prediction
The antigenic epitope within the oncogenic proteins (E6 and E7) and major capsid (L1) protein molecule of HPV-45 is predicted using ABCpred server (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html), a standard bioinformatics technique. All the parameters were in their default settings, but the epitopes selected had a score of more than 0.7. The ABCpred web server uses an artificial neural network to predict B-cell epitopes. This server is the first to use fixed-length patterns with a recurrent neural network (machine-based approach). This server can anticipate continuous (linear) B-cell epitopes. A linear B-cell epitope is a short peptide that binds to a conformational epitope and cross-reacts with an antibody. This server has a 65.93% accuracy rate in predicting epitopes.19,20
Protein structure analysis
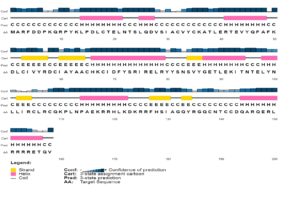
The E6, E7 and L1 proteins of HPV-45 contain 158, 106 and 539 amino acid residues and have a molecular weight of 18.89 kDa, 12.05 kDa and 60.31 kDa, respectively. ProtParam server was used to estimate the amino acid composition of all three proteins (Supplementary Table 1). The most common amino acids in E6 was arginine (R) (20 residues), followed by leucine (L) (15 residues). The secondary structure prediction revealed that 44.3% of the protein is coil (C), 41.8% is helix (H), and 13.9% is strand (E) (Figure 1). The most common amino acids in E7 protein was found to be leucine (L) (15 residues), followed by glutamic acid (E) (14 residues). The secondary structure prediction revealed that 65.1% of the protein is coil (C), 21.7% is helix (H), and 13.2% is strand (E) (Figure 2). The most common amino acids in L1 protein was proline (P), serine (S) and threonine (T) (42 residues), followed by valine (V) and leucine (L) (40 residues). The secondary structure prediction revealed that 63.5% of the protein is coil (C), 17.8% is helix (H), and 18.7% is strand (E). This was found to be the same in both the variant and reference sequences (Figure 3).
Prediction of epitopes for MHC-I alleles
IEDB server was used to predict epitopes for MHC-I alleles. It is crucial to understand the MHC-I and -II alleles that are highly expressed for the development of an efficient immunological response. The HLA allele reference set from the database and most frequently occurring MHC-I alleles were chosen for MHC-I binding. The immunogenicity score (<0.4) and percentile value (<0.5) were used to evaluate the possible epitopes (Table 1) for binding MHC-I alleles. HLA-A*01:01, HLA-A*02:01, HLA-A*02:03, HLA-A*02:06, HLA-B*07:02, HLA-B*15:01, HLA-A*30:02, HLA-A*31:01, HLA-A*33:01, HLA-B*35:01, HLA-B*40:01, HLA-B*44:03, HLA-B*57:01, HLA-B*58:01, HLA-A*68:01 are the highly expressed MHC-I alleles.
Table (1):
Predicted MHC-I epitopes for HPV-45 E6, E7 and L1 proteins through IEDB.
| No. | Start | End | Length | Peptide | Allele | Percentile Rank | Immunogenicity Score | Antigenicity |
|---|---|---|---|---|---|---|---|---|
| E6 | ||||||||
| 1 | 37 | 45 | 9 | ATLERTEVY | HLA-A*30:02, HLA-B*15:01, HLA-A*01:01, HLA-A*26:01, HLA-A*32:01, HLA-B*35:01, HLA-B*58:01, HLA-A*11:01 | 0.06-0.46 | 0.29271 | 0.6093 ( Probable ANTIGEN ) |
| 2 | 90 | 98 | 9 | LEKITNTEL | HLA-B*40:01 | 0.08 | 0.18689 | 0.4303 ( Probable ANTIGEN ) |
| 3 | 99 | 107 | 9 | YNLLIRCLR | HLA-A*33:01 | 0.34 | 0.11158 | 0.5910 ( Probable ANTIGEN ) |
| 4 | 70 | 78 | 9 | DFYSRIREL | HLA-B*08:01 | 0.05 | 0.11019 | 0.5919 ( Probable ANTIGEN ) |
| 5 | 122 | 130 | 9 | KDKRRFHSI | HLA-B*08:01 | 0.43 | 0.07332 | 0.9519 ( Probable ANTIGEN ) |
| 6 | 44 | 52 | 9 | VYQFAFKDL | HLA-A*24:02, HLA-A*23:01 | 0.2-0.27 | 0.05946 | 0.9584 ( Probable ANTIGEN ) |
| 7 | 13 | 21 | 9 | KLPDLCTEL | HLA-A*02:01, HLA-A*02:03, HLA-A*02:06 | 0.1-0.14 | 0.04843 | 0.9489 ( Probable ANTIGEN ) |
| 8 | 48 | 56 | 9 | AFKDLCIVY | HLA-A*30:02 | 0.15 | 0.02721 | 1.8184 ( Probable ANTIGEN ) |
| 9 | 113 | 121 | 9 | NPAEKRRHL | HLA-B*08:01, HLA-B*07:02 | 0.03-0.04 | 0.01475 | 0.9487 ( Probable ANTIGEN ) |
| E7 | ||||||||
| 10 | 19 | 27 | 9 | NELDPVDLL | HLA-B*40:01, HLA-B*44:03, HLA-B*44:02 | 0.05-0.26 | 0.05902 | 1.7631 ( Probable ANTIGEN ) |
| L1 | ||||||||
| 11 | 428 | 436 | 9 | ILENWNFGV | HLA-A*30:02 | 0.25 | 0.3542 | 1.1761 ( Probable ANTIGEN ) |
| 12 | 473 | 481 | 9 | KLKFWTVDL | HLA-A*32:01 | 0.29 | 0.34784 | 0.5705 ( Probable ANTIGEN ) |
| 13 | 17 | 25 | 9 | NVNVFPIFL | HLA-A*30:02 | 0.17 | 0.32372 | 0.5018 ( Probable ANTIGEN ) |
| 14 | 426 | 434 | 9 | SSILENWNF | HLA-A*30:02, HLA-A*32:01 | 0.19-0.4 | 0.30661 | 0.7650 ( Probable ANTIGEN ) |
| 15 | 409 | 417 | 9 | CTITLTAEV | HLA-A*30:02 | 0.08 | 0.19952 | 0.6661 ( Probable ANTIGEN ) |
| 16 | 19 | 27 | 9 | NVFPIFLQM | HLA-A*30:02, HLA-A*32:01, HLA-B*53:01, HLA-B*51:01 | 0.04-0.48 | 0.1896 | 0.8028 ( Probable ANTIGEN ) |
| 17 | 157 | 165 | 9 | ESAHAATAV | HLA-A*30:02 | 0.03 | 0.1758 | 0.5028 ( Probable ANTIGEN ) |
| 18 | 359 | 367 | 9 | FVTVVDTTR | HLA-A*30:02-0.42 | 0.19-0.42 | 0.17066 | 0.9193 ( Probable ANTIGEN ) |
| 19 | 271 | 279 | 9 | DSMFFCLRR | HLA-A*30:02 | 0.06-0.35 | 0.14493 | 0.8574 ( Probable ANTIGEN ) |
| 20 | 70 | 78 | 9 | TVGNPYFRV | HLA-A*30:02, HLA-A*02:06 | 0.13-0.5 | 0.11925 | 0.8608 ( Probable ANTIGEN ) |
| 21 | 499 | 507 | 9 | LVQAGLRRR | HLA-A*30:02 | 0.29 | 0.09825 | 1.1916 ( Probable ANTIGEN ) |
| 22 | 69 | 77 | 9 | LTVGNPYFR | HLA-A*68:01, HLA-A*30:02, HLA-A*33:01 | 0.05-0.21 | 0.09604 | 1.2652 ( Probable ANTIGEN ) |
| 23 | 67 | 75 | 9 | RLLTVGNPY | HLA-A*30:02, HLA-A*32:01 | 0.11-0.24 | 0.09562 | 0.5385 ( Probable ANTIGEN ) |
| 24 | 10 | 18 | 9 | GIIIFLKNV | HLA-A*02:03, HLA-A*02:06 | 0.45-0.5 | 0.0949 | 0.6342 ( Probable ANTIGEN ) |
| 25 | 274 | 282 | 9 | FFCLRREQL | HLA-B*08:01 | 0.22 | 0.08728 | 1.8410 ( Probable ANTIGEN ) |
| 26 | 398 | 406 | 9 | EEYDLQFIF | HLA-B*44:03, HLA-A*30:02 | 0.01-0.14 | 0.07784 | 1.7384 ( Probable ANTIGEN ) |
| 27 | 396 | 404 | 9 | HVEEYDLQF | HLA-B*35:01, HLA-B*53:01, HLA-A*26:01, HLA-A*01:01 | 0.24-0.47 | 0.07349 | 1.4499 ( Probable ANTIGEN ) |
| 28 | 132 | 140 | 9 | MEIGRGQPL | HLA-A*30:02, HLA-B*44:03, HLA-B*44:02 | 0.07-0.36 | 0.05536 | 0.9861 ( Probable ANTIGEN ) |
| 29 | 441 | 449 | 9 | TTSLVDTYR | HLA-A*68:01, HLA-A*33:01, HLA-A*31:01 | 0.02-0.41 | 0.02682 | 0.4094 ( Probable ANTIGEN ) |
| 30 | 452 | 460 | 9 | QSVAVTCQK | HLA-A*30:02 | 0.26-0.32 | 0.01633 | 1.3682 ( Probable ANTIGEN ) |
VaxiJen is the first server that allows antigen classification and predict protective antigens exclusively based on protein physicochemical properties rather than sequence alignment. Based on the high binding affinity score, the epitopes obtained from the IEDB server for MHC-I alleles were submitted to Vaxigen v2.0 for the prediction of probable antigens. The non-antigenic epitopes were removed and probable antigens were retained based on their antigenicity score.
Prediction of epitopes for MHC-II alleles
The epitopes for MHC-II alleles were predicted using IEDB server. The complete HLA reference set was chosen from the database for MHC-II binding. The potential epitopes (Table 2) for binding MHC-II alleles were assessed based on the percentile rank (<2.5). MHC-II alleles HLA-DRB1*01:01, HLA-DRB5*01:01, HLA-DRB3*02:02, HLA-DRB1*07:01, HLA-DRB1*09:01, HLA-DRB1*13:02, HLA-DRB1*15:01, HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*03:01/DPB1*04:02, HLA-DQA1*01:01/DQB1*05:01, HLA-DPA1*01:03/DPB1*04:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DPA1*02:01/DPB1*14:01, HLA-DQA1*05:01/DQB1*03:01, HLA-DPA1*02:01/DPB1*05:01 are majorly expressed. IFNepitope is an online prediction tool that seeks to predict and build peptides from protein sequences that can cause CD4+ T cells to release IFN-gamma. The MHC-II alleles retrieved from the IEDB server were further tested for IFN-g production, and those epitopes that were negative for IFN-g release were eliminated.
Table (2):
Predicted MHC-II epitopes for HPV-45 E6, E7 and L1 proteins through IEDB.
| No | Peptide | Length | Start | End | Allele | Percentile Rank | IFNg Score |
|---|---|---|---|---|---|---|---|
| E6 | |||||||
| 1 | SRIRELRYYSNSVYG | 15 | 73 | 87 | HLA-DRB1*15:01 | 0.49 | 1.000 |
| 2 | YSRIRELRYYSNSVY | 15 | 72 | 86 | HLA-DRB1*15:01 | 0.49 | 1.000 |
| 3 | IRELRYYSNSVYGET | 15 | 75 | 89 | HLA-DRB1*15:01 | 0.54 | 1.000 |
| 4 | RIRELRYYSNSVYGE | 15 | 74 | 88 | HLA-DRB1*15:01 | 0.54 | 1.000 |
| 5 | RELRYYSNSVYGETL | 15 | 76 | 90 | HLA-DRB1*15:01 | 1.3 | 1.000 |
| 6 | FHSIAGQYRGQCNTC | 15 | 127 | 141 | HLA-DRB5*01:01 | 1.6 | 0.151 |
| 7 | SNSVYGETLEKITNT | 15 | 82 | 96 | HLA-DPA1*03:01/DPB1*04:02 | 1.7 | 0.474 |
| 8 | YSNSVYGETLEKITN | 15 | 81 | 95 | HLA-DPA1*03:01/DPB1*04:02 | 1.7 | 0.146 |
| 9 | YYSNSVYGETLEKIT | 15 | 80 | 94 | HLA-DPA1*03:01/DPB1*04:02 | 1.7 | 0.489 |
| 15 | HLA-DPA1*02:01/DPB1*01:01 | 2.2 | 0.489 | ||||
| 10 | RTEVYQFAFKDLCIV | 15 | 41 | 55 | HLA-DQA1*01:01/DQB1*05:01 | 1.8 | 0.541 |
| 15 | HLA-DPA1*01:03/DPB1*04:01 | 2.1 | 0.541 | ||||
| 11 | TEVYQFAFKDLCIVY | 15 | 42 | 56 | HLA-DQA1*01:01/DQB1*05:01 | 1.8 | 0.660 |
| 15 | HLA-DPA1*01:03/DPB1*04:01 | 2.5 | 0.660 | ||||
| 12 | ELRYYSNSVYGETLE | 15 | 77 | 91 | HLA-DRB1*15:01 | 2.0 | 1.000 |
| 13 | RYYSNSVYGETLEKI | 15 | 79 | 93 | HLA-DPA1*02:01/DPB1*01:01 | 2.1 | 0.476 |
| 14 | ERTEVYQFAFKDLCI | 15 | 40 | 54 | HLA-DQA1*01:01/DQB1*05:01 | 2.5 | 0.205 |
| E7 | |||||||
| 15 | RTLQQLFLSTLSFVC | 15 | 85 | 99 | HLA-DPA1*01:03/DPB1*04:01 | 0.53 | 0.204 |
| 15 | HLA-DPA1*02:01/DPB1*01:01 | 1.5 | 0.204 | ||||
| 15 | HLA-DPA1*01:03/DPB1*02:01 | 2.0 | 0.204 | ||||
| 15 | HLA-DPA1*03:01/DPB1*04:02 | 2.3 | 0.204 | ||||
| 16 | LRTLQQLFLSTLSFV | 15 | 84 | 98 | HLA-DPA1*01:03/DPB1*04:01 | 0.61 | 0.264 |
| 15 | HLA-DPA1*02:01/DPB1*01:01 | 1.2 | 0.264 | ||||
| 15 | HLA-DPA1*01:03/DPB1*02:01 | 1.9 | 0.264 | ||||
| 15 | HLA-DPA1*03:01/DPB1*04:02 | 2.2 | 0.264 | ||||
| 17 | DLRTLQQLFLSTLSF | 15 | 83 | 97 | HLA-DPA1*01:03/DPB1*04:01 | 0.99 | 0.037 |
| 18 | QQLFLSTLSFVCPWC | 15 | 88 | 102 | HLA-DPA1*01:03/DPB1*04:01 | 1.2 | 0.045 |
| 15 | HLA-DPA1*01:03/DPB1*02:01 | 2.2 | 0.045 | ||||
| 19 | EDLRTLQQLFLSTLS | 15 | 82 | 96 | HLA-DPA1*01:03/DPB1*04:01 | 1.7 | 0.179 |
| 15 | HLA-DPA1*02:01/DPB1*01:01 | 2.0 | 0.179 | ||||
| 15 | HLA-DPA1*03:01/DPB1*04:02 | 2.3 | 0.179 | ||||
| 20 | AEDLRTLQQLFLSTL | 15 | 81 | 95 | HLA-DPA1*01:03/DPB1*04:01 | 1.8 | 0.214 |
| 15 | HLA-DPA1*02:01/DPB1*01:01 | 2.0 | 0.214 | ||||
| 15 | HLA-DPA1*03:01/DPB1*04:02 | 2.3 | 0.214 | ||||
| 21 | QLFLSTLSFVCPWCA | 15 | 89 | 103 | HLA-DPA1*01:03/DPB1*04:01 | 2.2 | 0.088 |
| L1 | |||||||
| 21 | AHNIIYGHGIIIFLK | 15 | 2 | 16 | HLA-DRB1*13:02 | 2 | 0.679 |
| 22 | AYQYRVFRVALPDPN | 15 | 94 | 108 | HLA-DPA1*02:01/DPB1*14:01 | 1.1 | 0.748 |
| 23 | DDTESAHAATAVITQ | 15 | 154 | 168 | HLA-DQA1*05:01/DQB1*03:01 | 1.5 | 0.66 |
| 24 | DTESAHAATAVITQD | 15 | 155 | 169 | HLA-DQA1*05:01/DQB1*03:01 | 1.5 | 0.843 |
| 15 | HLA-DQA1*01:02/DQB1*06:02 | 2.4 | 0.843 | ||||
| 25 | ESAHAATAVITQDVR | 15 | 157 | 171 | HLA-DQA1*01:02/DQB1*06:02 | 2.3 | 1.052 |
| 26 | FLKNVNVFPIFLQMA | 15 | 14 | 28 | HLA-DPA1*01:03/DPB1*04:01 | 2.5 | 0.178 |
| 27 | FLVQAGLRRRPTIGP | 15 | 498 | 512 | HLA-DRB5*01:01 | 1.1 | 0.151 |
| 28 | GRKFLVQAGLRRRPT | 15 | 495 | 509 | HLA-DRB5*01:01 | 0.13 | 0.615 |
| 15 | HLA-DPA1*02:01/DPB1*14:01 | 0.64 | 0.615 | ||||
| 29 | IFLKNVNVFPIFLQM | 15 | 13 | 27 | HLA-DRB3*02:02 | 1.3 | 0.137 |
| 15 | HLA-DPA1*01:03/DPB1*04:01 | 2.3 | 0.137 | ||||
| 15 | HLA-DRB1*13:02 | 2.3 | 0.137 | ||||
| 30 | IFYHAGSSRLLTVGN | 15 | 59 | 73 | HLA-DRB1*09:01 | 0.09 | 0.181 |
| 15 | HLA-DRB1*07:01 | 0.75 | 0.181 | ||||
| 15 | HLA-DRB1*01:01 | 1.2 | 0.181 | ||||
| 15 | HLA-DRB3*02:02 | 2.2 | 0.181 | ||||
| 31 | IIFLKNVNVFPIFLQ | 15 | 12 | 26 | HLA-DRB1*13:02 | 0.98 | 0.243 |
| 15 | HLA-DRB3*02:02 | 0.99 | 0.243 | ||||
| 15 | HLA-DRB1*15:01 | 2.5 | 0.243 | ||||
| 32 | KFLVQAGLRRRPTIG | 15 | 497 | 511 | HLA-DRB5*01:01 | 0.88 | 0.387 |
| 33 | KVSAYQYRVFRVALP | 15 | 91 | 105 | HLA-DPA1*01:03/DPB1*04:01 | 1.9 | 0.429 |
| 34 | LDDTESAHAATAVIT | 15 | 153 | 167 | HLA-DQA1*05:01/DQB1*03:01 | 2.5 | 0.58 |
| 35 | LGRKFLVQAGLRRRP | 15 | 494 | 508 | HLA-DRB5*01:01 | 0.13 | 0.386 |
| 15 | HLA-DPA1*02:01/DPB1*14:01 | 0.56 | 0.386 | ||||
| 36 | MAHNIIYGHGIIIFL | 15 | 1 | 15 | HLA-DRB1*13:02 | 1.8 | 0.55 |
| 37 | MFFCLRREQLFARHF | 15 | 273 | 287 | HLA-DPA1*02:01/DPB1*05:01 | 2.4 | 1 |
| 38 | PKVSAYQYRVFRVAL | 15 | 90 | 104 | HLA-DPA1*01:03/DPB1*04:01 | 2.2 | 0.775 |
| 39 | PLGRKFLVQAGLRRR | 15 | 493 | 507 | HLA-DRB5*01:01 | 0.13 | 0.371 |
| 15 | HLA-DPA1*02:01/DPB1*14:01 | 0.66 | 0.371 | ||||
| 40 | QYRVFRVALPDPNKF | 15 | 96 | 110 | HLA-DPA1*02:01/DPB1*14:01 | 0.6 | 0.797 |
| 41 | RHVEEYDLQFIFQLC | 15 | 395 | 409 | HLA-DQA1*01:01/DQB1*05:01 | 2 | 0.115 |
| 42 | RKFLVQAGLRRRPTI | 15 | 496 | 510 | HLA-DRB5*01:01 | 0.13 | 0.547 |
| 15 | HLA-DPA1*02:01/DPB1*14:01 | 1.2 | 0.547 | ||||
| 43 | RTSIFYHAGSSRLLT | 15 | 56 | 70 | HLA-DRB1*09:01 | 0.07 | 0.062 |
| 15 | HLA-DRB1*07:01 | 0.28 | 0.062 | ||||
| 15 | HLA-DRB1*01:01 | 0.67 | 0.062 | ||||
| 15 | HLA-DRB3*02:02 | 1.1 | 0.062 | ||||
| 15 | HLA-DRB5*01:01 | 2.2 | 0.062 | ||||
| 44 | SAYQYRVFRVALPDP | 15 | 93 | 107 | HLA-DPA1*02:01/DPB1*14:01 | 1.7 | 0.441 |
| 45 | SMFFCLRREQLFARH | 15 | 272 | 286 | HLA-DPA1*02:01/DPB1*05:01 | 2.5 | 1 |
| 46 | TESAHAATAVITQDV | 15 | 156 | 170 | HLA-DQA1*05:01/DQB1*03:01 | 1.8 | 0.957 |
| 15 | HLA-DQA1*01:02/DQB1*06:02 | 2.1 | 0.957 | ||||
| 47 | TSIFYHAGSSRLLTV | 15 | 57 | 71 | HLA-DRB1*09:01 | 0.07 | 0.063 |
| 15 | HLA-DRB1*07:01 | 0.28 | 0.063 | ||||
| 15 | HLA-DRB1*01:01 | 0.51 | 0.063 | ||||
| 15 | HLA-DRB3*02:02 | 1.1 | 0.063 | ||||
| 15 | HLA-DRB5*01:01 | 2.1 | 0.063 | ||||
| 48 | YPLGRKFLVQAGLRR | 15 | 492 | 506 | HLA-DRB5*01:01 | 0.13 | 0.368 |
| 15 | HLA-DPA1*02:01/DPB1*14:01 | 1.5 | 0.368 | ||||
| 49 | YQYRVFRVALPDPNK | 15 | 95 | 109 | HLA-DPA1*02:01/DPB1*14:01 | 0.66 | 1.049 |
| 50 | YRVFRVALPDPNKFG | 15 | 97 | 111 | HLA-DPA1*02:01/DPB1*14:01 | 1.4 | 1.122 |
Potential B-cell epitope prediction
The B-cell epitopes for HPV-45 E6, E7, and L1 proteins were predicted using the default settings of the ABCpred server (Table 3). B-cell epitopes are essential for cancer immunotherapy. In total, 12 potent B-epitopes were predicted for HPV-45 E6 protein. The most prominent epitope was SIAGQYRGQCNTCCDQ, with a binding score of 0.87. For HPV-45 E7 protein sequences, 6 potent B-epitopes were predicted, with the most prominent epitope LQEIVLHLEPQNELDP, with a binding score of 0.92. Whereas, for HPV-45 L1 protein sequences, 53 potent B-epitopes were predicted. The most prominent epitope was DSTVYLPPPSVARVVS, with a binding score of 0.96.
Table (3):
Predicted potential B-cell epitopes in HPV-45 E6, E7 and L1 proteins by ABCPred.
| Rank | Sequence | Start position | Score |
|---|---|---|---|
| E6 | |||
| 1 | SIAGQYRGQCNTCCDQ | 129 | 0.87 |
| 2 | YGETLEKITNTELYNL | 86 | 0.85 |
| 2 | SRIRELRYYSNSVYGE | 73 | 0.85 |
| 3 | HKCIDFYSRIRELRYY | 66 | 0.83 |
| 4 | MARFDDPKQRPYKLPD | 1 | 0.82 |
| 5 | CVYCKATLERTEVYQF | 32 | 0.81 |
| 5 | PDLCTELNTSLQDVSI | 15 | 0.81 |
| 6 | RRHLKDKRRFHSIAGQ | 118 | 0.80 |
| 7 | YNLLIRCLRCQKPLNP | 99 | 0.78 |
| 8 | QRPYKLPDLCTELNTS | 9 | 0.77 |
| 9 | YQFAFKDLCIVYRDCI | 45 | 0.74 |
| 10 | DCIAYAACHKCIDFYS | 58 | 0.72 |
| E7 | |||
| 1 | LQEIVLHLEPQNELDP | 8 | 0.92 |
| 2 | ADGVSHAQLPARRAEP | 42 | 0.89 |
| 3 | DGRIELTVESSAEDLR | 70 | 0.88 |
| 4 | AEPQRHKILCVCCKCD | 55 | 0.81 |
| 5 | ILCVCCKCDGRIELTV | 62 | 0.78 |
| 5 | SESEEENDEADGVSHA | 33 | 0.78 |
| L1 | |||
| 1 | DSTVYLPPPSVARVVS | 34 | 0.96 |
| 2 | VSAYQYRVFRVALPDP | 92 | 0.92 |
| 2 | AVTCQKDTTPPEKQDP | 455 | 0.92 |
| 2 | CQSICKYPDYLQMSAD | 252 | 0.92 |
| 3 | KFLVQAGLRRRPTIGP | 497 | 0.91 |
| 4 | RPTIGPRKRPAASTST | 507 | 0.89 |
| 4 | PPEKQDPYDKLKFWTV | 464 | 0.89 |
| 4 | DSTIYNPETQRLVWAC | 114 | 0.89 |
| 5 | LGCVPAIGEHWAKGTL | 186 | 0.88 |
| 5 | RVALPDPNKFGLPDST | 101 | 0.88 |
| 6 | RHVEEYDLQFIFQLCT | 395 | 0.87 |
| 6 | NGICWHNQLFVTVVDT | 350 | 0.87 |
| 7 | FWTVDLKEKFSSDLDQ | 476 | 0.86 |
| 7 | AHAATAVITQDVRDNV | 159 | 0.86 |
| 7 | LGIGLSGHPFYNKLDD | 140 | 0.86 |
| 7 | QRLVWACVGMEIGRGQ | 123 | 0.86 |
| 8 | SIITSDSQLFNKPYWL | 327 | 0.85 |
| 8 | GSCVYSPSPSGSIITS | 316 | 0.85 |
| 8 | ANMRETPGSCVYSPSP | 309 | 0.85 |
| 8 | RHFWNRAGVMGDTVPT | 285 | 0.85 |
| 8 | QMALWRPSDSTVYLPP | 26 | 0.85 |
| 8 | EHWAKGTLCKPAQLQP | 194 | 0.85 |
| 9 | TAEVMSYIHSMNSSIL | 414 | 0.84 |
| 9 | PVPSTYDPTKFKQYSR | 380 | 0.84 |
| 10 | GVMGDTVPTDLYIKGT | 292 | 0.83 |
| 11 | LQPGDCPPLELKNTII | 207 | 0.82 |
| 11 | QDVRDNVSVDYKQTQ | 167 | 0.82 |
| 12 | YFRVVPNGAGNKQAVP | 75 | 0.81 |
| 12 | YGHGIIIFLKNVNVFP | 7 | 0.81 |
| 12 | TSTASTASRPAKRVRI | 520 | 0.81 |
| 12 | TDLYIKGTSANMRETP | 300 | 0.81 |
| 12 | KNTIIEDGDMVDTGYG | 218 | 0.81 |
| 13 | TSLVDTYRFVQSVAVT | 442 | 0.80 |
| 14 | HSMNSSILENWNFGVP | 422 | 0.79 |
| 14 | DYLQMSADPYGDSMFF | 260 | 0.79 |
| 14 | GDMVDTGYGAMDFSTL | 225 | 0.79 |
| 14 | GMEIGRGQPLGIGLSG | 131 | 0.79 |
| 15 | LCTITLTAEVMSYIHS | 408 | 0.78 |
| 15 | TLCASTQNPVPSTYDP | 372 | 0.78 |
| 15 | SVDYKQTQLCILGCVP | 175 | 0.78 |
| 15 | PFYNKLDDTESAHAAT | 148 | 0.78 |
| 16 | SRTSIFYHAGSSRLLT | 55 | 0.77 |
| 17 | AGNKQAVPKVSAYQYR | 83 | 0.76 |
| 17 | SSDLDQYPLGRKFLVQ | 486 | 0.76 |
| 18 | TVVDTTRSTNLTLCAS | 361 | 0.75 |
| 18 | LHKAQGHNNGICWHNQ | 342 | 0.75 |
| 18 | STLQDTKCEVPLDICQ | 238 | 0.75 |
| 19 | VARVVSTDDYVSRTSI | 44 | 0.73 |
| 19 | YGDSMFFCLRREQLFA | 269 | 0.73 |
| 20 | RFVQSVAVTCQKDTTP | 449 | 0.72 |
| 21 | RREQLFARHFWNRAGV | 278 | 0.71 |
| 21 | YGAMDFSTLQDTKCEV | 232 | 0.71 |
| 22 | DPTKFKQYSRHVEEYD | 386 | 0.74 |
HPV-related cancers account for approximately 4.5% of all cancers, affecting nearly 600,000 people globally every year. Both E6 and E7 proteins of HPV promote excessive cell proliferation. E6 binds to and degrades p53 and other host cell proteins, whereas E7 binds to and degrades Retinoblastoma (Rb) protein. Both p53 and Rb protein are cellular growth repressors. HPV virions use L1 and L2 proteins to attach to the basal cells after infection. Antibodies bind to the virus, preventing infection by stopping it from infecting epithelial cells.21 An immunoglobulin-coated capsid is formed by high antibody titers, thus preventing the viral particle from attaching to basal cells, which is the initial stage of infection. As a result, neutrophils remove the virus that has been coated with antibodies. The virus particles are partially prevented from adhering to the basal cells in the presence of low antibody titers. The main mechanism of action is triggered by the capsid not binding to the second L1 receptor on the surface of the epithelial cell. As a result, the virus is removed from the tissue.22 E6, E7, and L1 proteins appear to be promising vaccine candidates due to the presence of numerous known neutralising epitopes.23
It is crucial to understand the vaccine candidate’s structural characteristics, such as its secondary structure. Alpha helix and coil-containing proteins and peptides are significant structural antigens because antibodies can identify them.24 The amino acid composition has shown that leucine residues are the most frequently occurring amino acids in all three proteins, i.e., E6, E7, and L1 of HPV-45. The most intriguing fact is that leucine residues have been studied earlier due to their significance in histone deacetylases (HDACs) binding. Histones attached to the MHC-I promoter are physically coupled with HDACs, and act as transcriptional co-repressors. It is likely that these HDACs cause MHC-I down-regulation due to the suppression of chromatin activation.25
In the present study, three potential B-cell epitopes were identified i.e., SIAGQYRGQCNTCCDQ, LQEIVLHLEPQNELDP, DSTVYLPPPSVARVVS, each in E6, E7 and L1 protein of HPV-45, respectively. The KLPDLCTEL epitope was predicted as a potential epitope for MHC class I alleles (HLA-A*02:01, HLA-A*02:03, HLA-A*02:06), similar to the HPV-18 validated epitopes,26,27 the predicted epitope can provide the cross-protection against HPV-18. Another potential epitope for E6 (ATLERTEVY) was predicted to 8 different MHC-I alleles (HLA-A*01:01, HLA-A*11:01, HLA-B*15:01, HLA-A*26:01, HLA-A*30:02, HLA-A*32:01, HLA-B*35:01, HLA-B*58:01) and for L1 epitope (NVFPIFLQM) was predicted for 4 MHC-I alleles (HLA-A*30:02, HLA-A*32:01, HLA-B*51:01, HLA-B*53:01).
As experimental procedures are labor-intensive and time-consuming, numerous in silico techniques for distinguishing protein epitopes are being developed. Computational techniques, on the other hand, provide quick, simple, cost-effective, and reliable methods for the prediction of immunogenic epitopes. Scientists can use bioinformatics tools to extract epitopes from a protein of interest instead of potential binding sites in epitope-based vaccinations. Moreover, enhanced computational model dependability for the prediction of desired epitopes will undoubtedly aid in the pre-experimental stage of vaccine development. Due to several limitations, such as the occurrence of diverse genotypes and vaccine price and accessibility, HPV prevention has remained a major problem. The most significant drawback appears to be the present vaccine’s limited coverage.28
A reference set of HLA alleles has been derived for both MHC I and MHC II binding prediction tools, providing more than 95% global population coverage, a significant characteristic for drug development. These techniques are useful for identifying a class of high-affinity binding peptides that could be produced and tested in the lab. The analysis projected the coverage of B and T cell epitope-based vaccinations in the population, allowing vaccines to be designed to maximise coverage.29
In silico approaches were used in this study to develop a vaccine candidate against oncoproteins and the major capsid protein of HPV-45. These proteins are strong candidates for antigenicity and immunogenicity due to their roles in viral replication, oncogenicity, and virus assembly. In this study, the amino acid sequence of the selected proteins was analysed, and their secondary structure was predicted. MHC-I and MHC-II epitopes for all three proteins were predicted and chosen based on their ability to induce antigenicity and produce IFN-γ, respectively. Further, B-cell epitopes were also predicted for the protein sequences. The epitopes identified by various web servers can be further used to create an effective antigenic vaccine capable of eliciting a significant immunological reaction over HPV-45. The discovery of potential epitopes has aided in developing cancer immunotherapy and detecting a wide range of infectious illnesses. Based on its rational design, we predict that the above-mentioned epitopes might be good candidates for vaccines against HPV-45 strains that are responsible for causing cervical cancer. It is possible to conduct additional molecular docking studies, followed by vaccine construct design using the predicted epitopes. It will require experimental confirmation by in vivo and in vitro studies, but it can be validated as a universally derived antigen when computationally analysed.
Additional file: Additional Table S1.
ACKNOWLEDGMENTS
The authors would like to thank Nitte (Deemed to be University) for providing a research facility.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PR and AK conceptualized the study. SP and AK applied methodology; SP wrote the original draft. PR, AK and BKK wrote, reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Qin F, Pang H, Yu T, Luo Y, Dong Y. Treatment strategies and prognostic factors of 2018 FIGO stage IIIC cervical cancer: a review. Technol Cancer Res Treat. 2022;21:15330338221086403.
Crossref - Tsang SH, Sampson JN, Schussler J, et al. Durability of cross-protection by different schedules of the bivalent HPV vaccine: the CVT trial. J Natl Cancer Inst. 2020;112(10):1030-1037.
Crossref - Dochez C, Bogers JJ, Verhelst R, Rees H. HPV vaccines to prevent cervical cancer and genital warts: an update. Vaccine. 2014;32(14):1595-601.
Crossref - Chen AA, Heideman DA, Boon D, et al. Human papillomavirus 45 genetic variation and cervical cancer risk worldwide. J Virol. 2014;88(8):4514-21.
Crossref - Namvar A, Panahi HA, Agi E, Bolhassani A. Development of HPV16, 18, 31, 45 E5 and E7 peptides-based vaccines predicted by immunoinformatics tools. Biotechnol Lett. 2020;42(3):403-418.
Crossref - Rezhake R, Hu SY, Zhao S, et al. Eight-type human papillomavirus E6/E7 oncoprotein detection as a novel and promising triage strategy for managing HPV-positive women. Int J Cancer. 2019;144(1):34-42.
Crossref - Bruno MT, Ferrara M, Fava V, Barrasso G, Panella MM. A prospective study of women with ASCUS or LSIL pap smears at baseline and HPV E6/E7 mRNA positive: a 3-year follow-up. Epidemiol Infect. 2018;146(5):612-618.}
Crossref - Kumar A, Yadav IS, Hussain S, Das BC, Bharadwaj M. Identification of immunotherapeutic epitope of E5 protein of human papillomavirus-16: An in silico approach. Biologicals. 2015;43(5):344-348.
Crossref - Smith JF, Brownlow M, Brown M, et al. Antibodies from Women Immunised with Gardasil ® Cross-Neutralize HPV 45 Pseudovirions. Hum Vaccin. 2007;3(4):109-115.
Crossref - Dehghani B, Hasanshahi Z, Hashempour T, Motamedifar M. The possible regions to design Human Papilloma Viruses vaccine in Iranian L1 protein. Biologicals. 2020;75(5):749-59.
Crossref - Pradini GW, Sahiratmadja E, Suhandono S, et al. Phylogeny and In Silico Structure Analysis of Major Capsid Protein (L1) Human Papillomavirus 45 from Indonesian Isolates. Asian Pac J Cancer Prev. 2020;21(9):2517.
Crossref - Qi W, Qingfeng L, Jing Z, et al. A novel multi-epitope vaccine of HPV16 E5 E6 E7 oncoprotein delivered by HBc VLPs induced efficient prophylactic and therapeutic antitumor immunity in tumor mice model. Vaccine. 2022;40(52):7693-702.
Crossref - Wang R, Pan W, Jin L, et al. Human papillomavirus vaccine against cervical cancer: Opportunity and challenge. Cancer Lett. 2020;471:88-102.
Crossref - Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. Proteom Protoc Handb. 2005:571-607.
Crossref - Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Bio. 1999;292(2):195-202.
Crossref - Vita R, Mahajan S, Overton JA, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339-43.
Crossref - Irini A Doytchinova and Darren R Flower. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4.
Crossref - Dhanda SK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8(1):1-5.
Crossref - Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40-48.
Crossref - Saha S, Raghava GP. Prediction methods for B-cell epitopes. Methods Mol Biol. 2007:387-394.
Crossref - Kesheh MM, Shavandi S, Jahromi ZK, Esghaei M, Keyvani H. Genetic diversity and bioinformatic analysis of the HPV-45 L1 gene in Iranian women with normal cytology. Hum Gene. 2022;34:201092.
Crossref - Rumfield CS, Roller N, Pellom ST, Schlom J, Jochems C. Therapeutic vaccines for HPV-associated malignancies. Immuno Targets and Therapy. 2020;9:167.
Crossref - Mahmoudvand S, Shokri S, Makvandi M, et al. In silico prediction of T-cell and B-cell epitopes of human papillomavirus type 16 L1 protein. Biotechnol Appl Biochem. 2021;69(2):514-525.
Crossref - Kumar A, Sahu U, Kumari P, Dixit A, Khare P. Designing of multi-epitope chimeric vaccine using immunoinformatic platform by targeting oncogenic strain HPV 16 and 18 against cervical cancer. Sci Rep. 2022;12(1):1-6.
Crossref - Heller C, Weisser T, Mueller-Schickert A, et al. Identification of key amino acid residues that determine the ability of high risk HPV16-E7 to dysregulate major histocompatibility complex class I expression. J Biol Chem. 2011;286(13):10983-10997.
Crossref - Matijevic M, Hedley ML, Urban RG, Chicz RM, Lajoie C, Luby TM. Immunisation with a poly (lactide co-glycolide) encapsulated plasmid DNA expressing antigenic regions of HPV 16 and 18 results in an increase in the precursor frequency of T cells that respond to epitopes from HPV 16, 18, 6 and 11. Cell Immunol. 2011;270(1):62-69.
Crossref - Panahi HA, Bolhassani A, Javadi G, Noormohammadi Z. A comprehensive in silico analysis for identification of therapeutic epitopes in HPV16, 18, 31 and 45 oncoproteins. PloS One. 2018;13(10):e0205933.
Crossref - Hosseini NG, Tebianian M, Farhadi A, et al. In silico analysis of L1/L2 sequences of human papillomaviruses: implication for universal vaccine design. Viral Immunol. 2017;30(3):210-23.
Crossref - Fleri W, Paul S, Dhanda SK, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.