ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is an important pathogen that is frequently associated with nosocomial infections. The goal of this work was to determine the relationship between the quorum sensing system (QS) and the production of virulence factors in P. aeruginosa. A number of 100 P. aeruginosa isolates were collected from various clinical sources from different Mansoura university hospitals in the period from April 2018 till April 2019. PCR screening of QS genes in the isolates was carried out including lasI, lasR, rhlI and rhlR. Thereafter, assay of the production of different virulence factors in the isolates was established including biofilm formation, pyocyanin production, protease production, lipase production, hemolysin production as well as swimming motility. Finally, statistical analysis of the data was performed to confirm the relationship between the QS and the production of virulence factors. Out of the 100 P. aeruginosa isolates, 27 clinical isolates were QS deficient. PCR analysis revealed that 8 isolates lacked lasR gene, 15 isolates lacked lasR and rhlR genes, 1 isolate lacked lasR and lasI genes, 2 isolates lacked lasR, lasI and rhlR genes and 1 isolate lacked rhlR, rhlI and lasR genes. There was a significant decrease observed in the production of pyocyanin, protease, lipase, hemolysin and biofilm formation as well as swimming motility in P. aeruginosa QS deficient isolates in comparison to non-QS deficient ones. There was a clear association between QS and virulence factors production in P. aeruginosa. This could open the door for novel promising targets for developing new therapeutic strategies against infections caused by this pathogen.
Pseudomonas aeruginosa, Quorum sensing, Regulation, Virulence factors
Pseudomonas aeruginosa is a Gram-negative rod-shaped bacterium, well known by its characteristic features as production of pyocyanin, oxidase positivity, grape-like odour and motility.1-3 P. aeruginosa is an opportunistic bacterium and it harbours a variety of virulence factors that are responsible for its danger.4 In hospitals, P. aeruginosa plays a pivotal role in nosocomial infections and is considered as a mutual cause of wound infections, especially for thermal burns.5
Community interaction between bacteria takes place via a well-developed framework called the quorum sensing system (QS).6,7 QS is a signalling process, which is used by different bacterial species to organize gene expression of the population according to changes in the cell density.1,8,9 QS is responsible for bacterial social behaviours, and this cell-to-cell communication or intercellular signalling takes place in Gram-negative bacteria via small N- acylated homoserine lactone molecules called autoinducers (AIs). These AIs coordinate the common actions associated with the succession of bacterial infection, which depends on the expression of virulence factors and invasion abilities.1,8,10,11
P. aeruginosa QS network is structured in a multi-layered hierarchy manner composed of at least four linked signalling systems, which are Las, Rhl, PQS and IQS.3,8,10,12 Among these systems, the two well-defined interconnected QS systems, Las and Rhl, are well defined to control virulence factors production.1,8,10,12 Other functions have been correlated with QS systems, which are involved in cell metabolism, stress responses, etc.8 There is clear evidence that the QS regulatory network not only reacts to changes in the bacterial population, but can also react to environmental changes.13,14 Thus, it is well established that during the discovery and development of anti-QS therapeutics, this plasticity should be taken into account.9
Clinical studies have shown that QS systems in infected tissues are fully functional, especially in patients with cystic fibrosis (CF) lungs who have been chronically infected with P. aeruginosa.15-19 The sputum analysis of P. aeruginosa infection in CF patients revealed the existence of transcripts for both lasI and lasR.15,18 Accumulation of these transcripts was associated with cumulation of transcripts for the lasB, lasA and toxA QS-controlled genes.15,18 In addition, the presence of AIs within the sputum of CF patients was confirmed in other studies.20,21 Moreover, the biological activation of these AIs was proved in other reports.15
In numerous P. aeruginosa infected animal models, the importance of QS in regulating virulence has been described involving the thermally injured mouse model as well as mouse models of acute and chronic lung infections.22-26 Mutants of P. aeruginosa with deletions in the QS genes were compared to their parent strains according to their virulence. These studies showed that mice infected with QS mutants and thermally injured have lower mortality rate than mice infected with the parent strain.23,24 The spreading of the mutant strains decreased locally in the skin of thermally damaged mice and decreased systemically in the infected thermally injured mice bodies.23,24 Using the P. aeruginosa acute pulmonary infection mouse model, it was demonstrated that the mortality rate and the lung damage decreased with P. aeruginosa strains that carry deletions within the QS genes.22 In addition, some studies reported that flat biofilms are produced by lasI deficient strains in which this produced biofilm is different from the normal wild types.27 Another report indicated that QS deficiency takes place naturally and Las QS has important role in corneal infection development.28 Mice models that were infected by mutants deficient in the lasI gene exhibited virulence reduction, and by implantation of functional lasI gene, lasI mutant virulence was fully restored.29 From this background, in this study, we were motivated to screen the presence of QS genes in P. aeruginosa clinical isolates collected from different sources in Mansoura university hospitals, Egypt, and to reveal the impact of QS on virulence factors production in this pathogen.
Bacterial isolates
This work was established after the agreement of the research ethics committee of the Faculty of Pharmacy, Tanta University, Egypt (Research Ethics Committee Code: TP / RE /12-21-M-002). A total of 100 P. aeruginosa clinical isolates were collected from different clinical sources including 35 from urine (U), 17 from wounds (W), 15 from burns (B), 14 from sputum (S), 10 from ear (E) and 9 from eye (EY). These isolates were collected from patients in different Mansoura hospitals under medical attention with strict aseptic precautions in the period from April 2018 till April 2019. According to the standard microbiological techniques,30 the isolates were biochemically identified as P. aeruginosa. Moreover, identity of the isolates was further confirmed by PCR amplification of their 16s rRNA as previously described.31 All cultures were grown at 37°C in Luria Bertani medium (LB broth; tryptone 1% w/v, yeast extract 0.5% w/v, and NaCl 1.0% w/v), otherwise specified, and stored in 80% glycerol/LB broth at -80°C.
Polymerase chain reaction
Chromosomal DNA was extracted from P. aeruginosa isolates by using QIA amp® DNA miniprep kit (Qiagen, Germany). All P. aeruginosa clinical isolates were screened for the presence of QS genes by PCR using the primers (lasR, lasI, rhlR and rhlI) shown in (Table 1) as previously reported.32,33 The volume of PCR reactions was 25 μL, and each reaction composed of 12.5 μL Dream Taq PCR master mix 2x (Fermentas, USA), 1 μL forward primer (10 μM), 1 μL of reverse primer (10 μM) and 3 μL of template DNA, then the reaction was adjusted to total volume of 25 μL with nuclease free water. Negative control tubes were also performed without template DNA. The cycling conditions performed were; initial denaturing at 95°C for 5 min, then 35 cycles of (denaturation at 95°C for 30 s, annealing at 50°C for 30 s and extension at 72°C for 2 min), and final extension at 72°C for 10 min. Visualization of the amplified genes by electrophoresis was done using 1% agarose gel stained with ethidium bromide (MP biomedicals, France), and compared with a 100 base pair plus DNA ladder (Thermo Scientific, UK). The presence of the tested gene was indicated by the appearance of a single sharp band with the specific amplicon size for each gene (Table 1).
Table (1):
Specific amplification primer sets for the tested quorum sensing genes among P. aeruginosa isolates.
| Gene name | Type | Sequence | Amplicon size (bp) | Annealing temperature |
Reference |
|---|---|---|---|---|---|
|
lasR |
F | ATGGCCTTGGTTGACGGTT | 725 | 50°C | 32, 33 |
| R | GCAAGATCAGAGAGTAATAAGACCCA | ||||
|
lasI |
F | ATGATCGTACAAATTGGTCGGC | 605 | 50°C | 32, 33 |
| R | GTCATGAAACCGCCAGTCG | ||||
|
rhlR |
F | CAATGAGGAATGACGGAGGC | 730 | 50°C | 32, 33 |
| R | GCTTCAGATGAGGCCCAGC | ||||
|
rhlI |
F | CTTGGTCATGATCGAATTGCTC | 625 | 50°C | 32, 33 |
| R | ACGGCTGACGACCTCACAC |
F: Forward; R: Reverse
Assay of P. aeruginosa virulence factors
Pyocyanin assay
Pyocyanin was extracted as previously described.34 King’s A liquid medium (peptone 2% w/v, K2SO4 1.0% w/v, and MgCl2 0.14% w/v) was used to cultivate the isolates for 48 hours with shaking at 150 rpm. Cultures were then centrifuged and pyocyanin was recovered from the supernatant using 3 mL chloroform. Thereafter, 1 mL of 0.2 N HCl was added to the pyocyanin (bottom layer) after it was transferred to a new clean tube. The extracted acidic form of pyocyanin, which has a pink colour, was spectrophotometrically measured at OD520 nm. The optical density at 520 nm was multiplied by 17.072 to get the microgram amounts of pyocyanin.35 All P. aeruginosa isolates were compared with the isolate with the highest amount of pyocyanin in percentage. In this experiment, King’s A liquid medium was used as a negative control.
Hemolysin assay
P. aeruginosa clinical isolates were cultured overnight in LB and incubated at 37°C with shaking at 150 rpm for an appropriate time. Thereafter, cultures were centrifuged at 10,000 rpm for 10 min at 4°C. The obtained supernatants were filtered through 0.2 µM Millipore filter and subsequently used in the assay of hemolysin activity. The assay was performed as previously described.36 Prepared free supernatant of bacterial cultures (volume 600 µL) was mixed with equal volume of saline suspension of erythrocytes 2%, and incubated at 37°C for 2 hours. Then, centrifugation of the reaction mixtures was established at 10,000 rpm for 8 min. at 4°C, and the optical density at 540 nm was used to determine the degree of haemoglobin release. Control studies for spontaneous (negative control) and complete (positive control) lysis were conducted without hemolysin and with 0.2 % sodium dodecyl sulphate, respectively. The percentage (%) of cells lysed = [(X-B)/ (T-B)] × 100, where B (baseline) is a negative control and T is a positive control corresponding to the total lysis.
Protease assay
Assay of total proteases was established using skimmed milk assay as previously illustrated.37 Volume of 200 µL of P. aeruginosa supernatant (prepared as described above in hemolysin assay) was mixed with 1 mL 1.25% skimmed milk and at 37°C incubation takes place for 15 min, and then, OD600 was measured. Comparison of OD600 of all P. aeruginosa isolates with OD600 of skimmed milk in percentage was established to detect the protease activity of each isolate. Protease activity = (OD600 of skimmed milk – OD600 of the sample) X 100. In this experiment, LB was used as a negative control.
Lipase assay
Lipase assay was established as formerly outlined.38 Culture supernatants of P. aeruginosa isolates were prepared as described above in hemolysin assay. A stock solution of p-nitrophenyl palmitate (p-NPP) (Sigma, USA) was prepared in HPLC grade of isopropanol. The reaction mixture contained 75 µL of p-NPP stock solution, 5 µL bacterial supernatant, and completed to a final volume of 3 mL with 0.1 M Tris buffer (pH 8.5). To stop the reaction, the reaction mixture was incubated at 37°C for 10 min before being refrigerated at -20°C for 8 min. At OD410 nm, the optical density of emitted p-nitrophenol was measured. All P. aeruginosa isolates were compared with the isolate with the highest lipase activity in percentage. In this experiment, LB was used as a negative control.
Swimming motility assay
Swarming motility assay was performed as previously depicted.39 Surface inoculation was used to measure the swimming ability of P. aeruginosa by using swimming agar plates (tryptone 1%, sodium chloride 0.5%, and agar 0.3%). The prepared plates were centrally stabbed with 5 μL of diluted overnight culture of P. aeruginosa isolates in tryptone broth. After 24 hours incubation period at 37°C, the swimming zones were measured. All P. aeruginosa isolates were compared with the isolate with the highest swimming activity in percentage. In this experiment, LB was used as a negative control.
Quantitative detection of biofilm using microtiter plate assay
By using the method described previously,40,41 biofilm production was assayed. P. aeruginosa clinical isolates were cultured overnight in tryptic soy broth (TSB) (Oxoid, Thermo Fisher, UK) and diluted to produce a cell density of approximately 1 × 106 CFU/mL. Using 96-well flat-bottomed polystyrene microtiter plate, 100 μL aliquots of each bacterial suspension was inoculated into the wells of at least in triplicate. Then, plates were incubated at 37°C for 24 hours. The contents of each well were aspirated to eliminate all non-adherent cells. Each well was rinsed three times with phosphate buffer saline (PBS, pH 7.4). The remaining adherent bacteria were fixed with 100% methanol per well. The bacterial cells adhered to the well walls were stained with 1% crystal violet for 20 min. Thereafter, the dye bound to the adhering cells was resolubilized using 33% (v/v) glacial acetic acid. The absorbance of solubilized stain was measured at OD492 nm using Biotek spectrofluorimeter (Biotek, USA). All P. aeruginosa isolates were compared with the isolate with the highest biofilm formation in percentage. In this experiment, LB was used as a negative control.
Statistical analysis
Significance of difference was assessed in the results by repeating the experiments three independent times and comparing the data using Student’s t-test. P values lower than 0.05 were regarded significantly different.
Detecting the presence of QS-genes in P. aeruginosa isolates
In this study there were 27 isolates out of the 100 P. aeruginosa clinical isolates were QS deficient isolates (Table 2). PCR analysis revealed that 8 isolates lacked lasR gene, 15 isolates lacked lasR and rhlR genes, 1 isolate lacked lasR and lasI genes, 2 isolates lacked lasR, lasI and rhlR genes and 1 isolate lacked rhlR, rhlI and lasR genes. Among the QS deficient P. aeruginosa isolates, 8 were collected from urine, 6 isolates from wounds, 6 isolates from burns, 3 isolates from sputum, 2 isolates from ear and 2 isolates from eye.
Assay of P. aeruginosa virulence factors
Pyocyanin assay
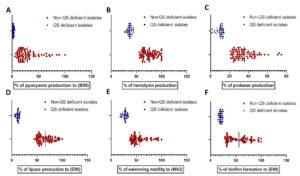
As shown in (Table 2), percentages of pyocyanin produced by P. aeruginosa QS deficient isolates according to the sample (B59), which produced the highest amount of pyocyanin, were in between 4.8% and 0.098%. Sample W50 had the lowest pyocyanin production percentage of 5.57% in all non-QS deficient isolates. The amount of pyocyanin production that was produced by P. aeruginosa QS deficient isolates was less than the isolates that possess all QS genes (Fig. 1A).
Fig. 1. Virulence factors production and biofilm formation in P. aeruginosa non-QS deficient and QS deficient isolates. (A) Pyocyanin production. (B) Hemolysin production. (C) Protease production. (D) Lipase production. (E) Swimming motility. (F) Biofilm formation.
Hemolysin assay
As illustrated in (Table 2), a number of 73 non-QS deficient isolates produced more than 50% hemolysis including 6 isolates that produced complete (100%) hemolysis. On the other hand, the 27 QS deficient isolates produced less than 50% hemolysis (Fig. 1B). This gives indication that QS deficient isolates produce hemolysin less than non-QS deficient isolates.
Protease assay
As depicted in (Table 2), the activity of QS deficient isolates was in between 14% and 1.3%, while isolate B64 had the lowest protease activity (21%) in all non-QS deficient isolates. Skimmed milk assay indicates that QS deficient isolates have lower protease activity in comparison to other non-QS deficient isolates (Fig. 1C).
Lipase assay
As demonstrated in (Table 2), percentage of lipase activity produced by P. aeruginosa isolates according to the sample (E90), which had the highest lipase activity, was in the range between 16.61% and 6.1%. Sample S72 had the lowest lipase activity percentage (44.37%) in all non-QS deficient isolates. The amount of lipase produced by QS deficient isolates is less in comparison to the other isolates, which possess all QS genes that were examined in this study (Fig. 1D).
Swimming motility assay
As depicted in (Table 2), percentages of swimming zone diameters produced by P. aeruginosa QS deficient isolates according to the sample (W43), which had the highest swimming zone diameter, were in the range between 34.375% and 18.125%. Sample U23 had the lowest swimming zone diameter percentage (36.875%) in all non-QS deficient isolates in this study (Fig. 1E). This gives an indication that the swimming motility of QS deficient isolates is less than the non-QS deficient ones.
Quantitative assay of biofilm formation
As shown in (Table 2), 24 QS deficient isolates were characterized by weak biofilm formation. Moreover, three isolates that lacked Las system (lasI and lasR genes were absent) were non-adherent. Percentage of biofilm formation produced by QS deficient isolates according to sample (E88), which had the highest capability of biofilm formation, was in the range between 25.17% and 14.12%. Sample U14 was weakly adherent and had the lowest biofilm formation percentage (28%) in all non-QS deficient isolates. The amount of biofilm formation, which produced by QS deficient isolates is less than the other of non-QS deficient isolates (Fig. 1F).
Table (2):
Results of tested quorum sensing genes among P. aeruginosa isolates and their impact on different phenotypes.
Isolate code |
Isolate source |
lasR |
lasI |
rhlR |
rhlI |
% of pyocyanin production to (B59) |
% of hemolysin production |
% of protease production |
% of lipase production to (E90) |
% of swimming motility to (W43) |
% of biofilm formation to (E88) |
|---|---|---|---|---|---|---|---|---|---|---|---|
U1 |
Urine |
+ |
+ |
+ |
+ |
20.88 |
70.12 |
23 |
52.24 |
48.125 |
67.5 |
U2 |
Urine |
+ |
+ |
+ |
+ |
20.34 |
73.57 |
30 |
59.45 |
60.625 |
63.48 |
U3 |
Urine |
+ |
+ |
+ |
+ |
20 |
62.29 |
35 |
51.03 |
46.25 |
66.66 |
U4 |
Urine |
+ |
+ |
+ |
+ |
18.9 |
79.57 |
26 |
68.19 |
44.6875 |
30.44 |
U5 |
Urine |
+ |
+ |
+ |
+ |
25.97 |
83.13 |
43.3 |
76.93 |
38.125 |
46.24 |
U6 |
Urine |
+ |
+ |
+ |
+ |
26.04 |
64.83 |
41.6 |
57.37 |
47.5 |
60.73 |
U7 |
Urine |
+ |
+ |
+ |
+ |
24.63 |
78.048 |
38.33 |
86 |
43.75 |
81.37 |
U8 |
Urine |
+ |
+ |
+ |
+ |
20.2 |
89.63 |
30.3 |
58.14 |
48.125 |
33.69 |
S9 |
Sputum |
+ |
+ |
+ |
+ |
23.6 |
59.75 |
45.6 |
73.77 |
43.125 |
64.61 |
U10 |
Urine |
+ |
+ |
+ |
+ |
19 |
67 |
21.3 |
52.67 |
48.125 |
87.81 |
U11 |
Urine |
+ |
+ |
+ |
+ |
40.18 |
91.56 |
44.6 |
78.68 |
56.875 |
80.90 |
U12 |
Urine |
+ |
+ |
+ |
+ |
20.53 |
96.44 |
57 |
49 |
48.125 |
80.31 |
U13 |
Urine |
+ |
+ |
+ |
+ |
12.76 |
61.58 |
27 |
85.57 |
48.75 |
39.37 |
U14 |
Urine |
+ |
+ |
+ |
+ |
13.77 |
78.45 |
31 |
69.94 |
46.25 |
28 |
S15 |
Sputum |
+ |
+ |
+ |
+ |
15.6 |
73.67 |
24.3 |
58.57 |
38.125 |
82.87 |
EY16 |
Eye |
+ |
+ |
+ |
+ |
32.11 |
66.56 |
46.6 |
78.14 |
65 |
50.75 |
EY17 |
Eye |
+ |
+ |
+ |
+ |
17.35 |
62.093 |
36.6 |
54.42 |
59.375 |
35 |
EY18 |
Eye |
+ |
+ |
+ |
+ |
25.2 |
62.60 |
32.6 |
76 |
46.875 |
40.57 |
EY19 |
Eye |
+ |
+ |
+ |
+ |
14.16 |
97.45 |
53.3 |
54.64 |
40.625 |
30.99 |
EY20 |
Eye |
+ |
+ |
+ |
+ |
12 |
93.90 |
44 |
62.73 |
46.25 |
32.45 |
EY21 |
Eye |
+ |
+ |
+ |
+ |
69.8 |
82.11 |
35 |
55.73 |
46.25 |
33.40 |
EY22 |
Eye |
+ |
+ |
+ |
+ |
29.7 |
71.95 |
33 |
72.67 |
71.25 |
43 |
U23 |
Urine |
+ |
+ |
+ |
+ |
14.9 |
62.70 |
23.6 |
63.49 |
36.875 |
39.37 |
U24 |
Urine |
+ |
+ |
+ |
+ |
18.63 |
58.73 |
39.6 |
53.66 |
57.5 |
48.84 |
U25 |
Urine |
+ |
+ |
+ |
+ |
50.53 |
100 |
71 |
51.80 |
45.625 |
94.47 |
U26 |
Urine |
+ |
+ |
+ |
+ |
10.93 |
83.63 |
35.67 |
76.93 |
70 |
66.55 |
U27 |
Urine |
+ |
+ |
+ |
+ |
25.30 |
75.20 |
43 |
93.87 |
45.625 |
49.87 |
U28 |
Urine |
+ |
+ |
+ |
+ |
60.15 |
64.12 |
74.6 |
84 |
80 |
41.74 |
U29 |
Urine |
+ |
+ |
+ |
+ |
29.96 |
87.70 |
41.6 |
95.3 |
40 |
99.89 |
S30 |
Sputum |
+ |
+ |
+ |
+ |
49.88 |
94.71 |
61 |
89 |
90.625 |
81.22 |
U31 |
Urine |
+ |
+ |
+ |
+ |
40.70 |
99.49 |
35.3 |
93.22 |
71.25 |
78.9 |
U32 |
Urine |
+ |
+ |
+ |
+ |
32.58 |
100 |
72.3 |
62.29 |
48.75 |
72.37 |
S33 |
Sputum |
+ |
+ |
+ |
+ |
14.14 |
81.70 |
30.3 |
90.81 |
41.25 |
28.39 |
U34 |
Urine |
+ |
+ |
+ |
+ |
31.39 |
75 |
32 |
54.20 |
43.125 |
32.12 |
U35 |
Urine |
– |
+ |
+ |
+ |
3.5 |
44.61 |
14 |
14.97 |
34.375 |
23.38 |
U36 |
Urine |
+ |
+ |
+ |
+ |
22.26 |
82.62 |
34.3 |
49 |
77.5 |
33.18 |
U37 |
Urine |
+ |
+ |
+ |
+ |
34.09 |
67.68 |
34.3 |
86.12 |
45.625 |
80.20 |
U38 |
Urine |
+ |
+ |
+ |
+ |
9.89 |
71.74 |
51.6 |
72.67 |
45.625 |
32.19 |
U39 |
Urine |
+ |
+ |
+ |
+ |
55.68 |
86.38 |
23.6 |
62.62 |
93.125 |
30 |
U40 |
Urine |
– |
+ |
+ |
+ |
3.85 |
44.7 |
13.6 |
15.62 |
29.375 |
22.64 |
W41 |
wounds |
+ |
+ |
+ |
+ |
13.9 |
56.40 |
45.6 |
52.45 |
56.875 |
31.75 |
W42 |
wounds |
+ |
+ |
+ |
+ |
10.86 |
61.788 |
28 |
57.15 |
84.375 |
36.99 |
W43 |
wounds |
+ |
+ |
+ |
+ |
6.813 |
62.70 |
29.6 |
49.61 |
100 |
95.31 |
W44 |
wounds |
+ |
+ |
+ |
+ |
16.21 |
72.76 |
22.3 |
49 |
93.125 |
43.61 |
W45 |
wounds |
– |
+ |
+ |
+ |
3.92 |
39.93 |
13.3 |
15.62 |
32.5 |
23.81 |
W46 |
wounds |
+ |
+ |
+ |
+ |
24.43 |
100 |
23 |
50.81 |
55 |
32.12 |
W47 |
wounds |
– |
+ |
+ |
+ |
4.2 |
43.5 |
12.6 |
13.33 |
31.25 |
23.38 |
W48 |
wounds |
– |
+ |
+ |
+ |
3.6 |
39.73 |
12 |
14.97 |
30.625 |
24.44 |
W49 |
wounds |
– |
+ |
+ |
+ |
2.041 |
39.6 |
13.3 |
12.7 |
33.125 |
23.45 |
W50 |
wounds |
+ |
+ |
+ |
+ |
5.57 |
86.48 |
33.3 |
52.89 |
40.625 |
58.28 |
W51 |
wounds |
+ |
+ |
+ |
+ |
64.99 |
65.04 |
24.6 |
72.67 |
53.75 |
32.12 |
W52 |
wounds |
+ |
+ |
+ |
+ |
22.51 |
91.66 |
33.3 |
66.33 |
61.25 |
35.12 |
W53 |
wounds |
+ |
+ |
+ |
+ |
10.44 |
84.75 |
39.3 |
80.21 |
40 |
39.59 |
W54 |
wounds |
+ |
+ |
+ |
+ |
40.21 |
59.55 |
29 |
58.79 |
45 |
30.58 |
W55 |
wounds |
+ |
+ |
+ |
+ |
17.57 |
67.88 |
38.6 |
52 |
49.375 |
38.74 |
B56 |
Burns |
+ |
+ |
+ |
+ |
84.2 |
100 |
46.3 |
65.35 |
87.5 |
69.48 |
B57 |
Burns |
+ |
+ |
+ |
+ |
12.21 |
64.43 |
25 |
77 |
70 |
32.38 |
B58 |
Burns |
+ |
+ |
+ |
+ |
31.81 |
73.78 |
51.3 |
80 |
56.25 |
60.88 |
B59 |
Burns |
+ |
+ |
+ |
+ |
100 |
100 |
32.6 |
91.47 |
58.75 |
77.71 |
B60 |
Burns |
+ |
+ |
+ |
+ |
20.39 |
55.79 |
40.6 |
47.54 |
40.625 |
31.72 |
B61 |
Burns |
+ |
+ |
+ |
+ |
12.811 |
60.87 |
46.3 |
55.3 |
45.625 |
35 |
B62 |
Burns |
+ |
+ |
+ |
+ |
12.04 |
63.414 |
28 |
74.53 |
40.625 |
52.50 |
B63 |
Burns |
+ |
+ |
+ |
+ |
26.01 |
74.69 |
29.6 |
74.53 |
54.375 |
70.25 |
B64 |
Burns |
+ |
+ |
+ |
+ |
75.31 |
75.30 |
21 |
55.95 |
38.75 |
41.93 |
B65 |
Burns |
– |
– |
+ |
+ |
0.57 |
31.7 |
4.6 |
8.41 |
20 |
15.18 |
S66 |
Sputum |
+ |
+ |
+ |
+ |
34.06 |
63.92 |
29 |
55.62 |
58.125 |
65.20 |
S67 |
Sputum |
+ |
+ |
+ |
+ |
17.72 |
86.38 |
27.3 |
54.31 |
50 |
33.15 |
S68 |
Sputum |
+ |
+ |
+ |
+ |
9.8 |
71.036 |
26.6 |
68.08 |
93.125 |
32.30 |
S69 |
Sputum |
+ |
+ |
+ |
+ |
28.23 |
65.85 |
31 |
49.18 |
80 |
68.31 |
S70 |
Sputum |
+ |
+ |
+ |
+ |
64.60 |
83.84 |
24.6 |
54.20 |
89.375 |
63.59 |
S71 |
Sputum |
+ |
+ |
+ |
+ |
14.735 |
56.30 |
30.3 |
69.39 |
42.5 |
63.11 |
S72 |
Sputum |
+ |
+ |
+ |
+ |
71.21 |
58.73 |
21.3 |
44.37 |
46.25 |
94.25 |
B73 |
Burns |
– |
+ |
– |
+ |
1.6 |
35 |
11.6 |
10.81 |
22.5 |
18.47 |
W74 |
Wounds |
– |
+ |
– |
+ |
1.03 |
37 |
10.3 |
12 |
26.25 |
19 |
B75 |
Burns |
– |
+ |
– |
+ |
1.75 |
35.6 |
12 |
8.9 |
25.625 |
17 |
U76 |
Urine |
– |
+ |
– |
+ |
1.75 |
36.1 |
9 |
11.91 |
28.125 |
21.62 |
S77 |
Sputum |
– |
+ |
– |
+ |
1.03 |
37 |
12.3 |
13.22 |
28.125 |
21.51 |
U78 |
Urine |
– |
+ |
– |
+ |
1.85 |
33.8 |
9.6 |
12.02 |
26.25 |
17.34 |
S79 |
Sputum |
– |
+ |
– |
+ |
1.48 |
36.99 |
10.3 |
11.25 |
26.875 |
17.52 |
U80 |
Urine |
– |
+ |
– |
+ |
1.23 |
35.1 |
7.3 |
12.67 |
28.125 |
16.79 |
EY81 |
Eye |
– |
+ |
– |
+ |
2.07 |
33.84 |
11 |
12.56 |
28.125 |
20.23 |
S82 |
Sputum |
– |
+ |
– |
+ |
2.17 |
37.5 |
9.6 |
13.33 |
26.875 |
17.70 |
EY83 |
Eye |
– |
+ |
– |
+ |
1.87 |
34.65 |
10 |
10.2 |
27.5 |
18.25 |
B84 |
Burns |
– |
+ |
– |
– |
0.28 |
30.5 |
4 |
6.77 |
20.625 |
19.1 |
U85 |
Urine |
– |
– |
– |
+ |
0.27 |
29.1 |
1.6 |
6.44 |
18.75 |
14.96 |
U86 |
Urine |
– |
– |
– |
+ |
0.098 |
26.72 |
1.3 |
6.1 |
18.125 |
14.12 |
E87 |
Ear |
+ |
+ |
+ |
+ |
8.56 |
93.39 |
28.3 |
60.43 |
40.625 |
85.72 |
E88 |
Ear |
+ |
+ |
+ |
+ |
33.25 |
65.54 |
23 |
81.20 |
51.25 |
100 |
E89 |
Ear |
+ |
+ |
+ |
+ |
43.74 |
77.13 |
26.3 |
56.83 |
40 |
65.78 |
E90 |
Ear |
+ |
+ |
+ |
+ |
88.86 |
100 |
33.6 |
100 |
58.75 |
66.70 |
E91 |
Ear |
+ |
+ |
+ |
+ |
15.84 |
72.25 |
24.3 |
74.97 |
53.75 |
95.35 |
E92 |
Ear |
+ |
+ |
+ |
+ |
18.78 |
85.67 |
49.3 |
87.21 |
66.875 |
80.75 |
E93 |
Ear |
+ |
+ |
+ |
+ |
16.29 |
88.61 |
25 |
53.11 |
49.375 |
64.58 |
E94 |
Ear |
+ |
+ |
+ |
+ |
41.15 |
81.80 |
28.6 |
71.36 |
79.375 |
72.63 |
B95 |
Burns |
– |
+ |
+ |
+ |
3.85 |
40.4 |
12.3 |
14.97 |
33.125 |
22.17 |
W96 |
Wounds |
– |
+ |
– |
+ |
2.44 |
38 |
10.3 |
11.7 |
28.75 |
18 |
E97 |
Ear |
– |
+ |
+ |
+ |
4.8 |
41.56 |
12 |
16.61 |
31.25 |
25.17 |
U98 |
Urine |
– |
+ |
– |
+ |
2.36 |
34.6 |
11.3 |
12.15 |
28.75 |
19.46 |
E99 |
Ear |
– |
+ |
– |
+ |
1.65 |
36.58 |
11.6 |
11.03 |
28.75 |
20 |
B100 |
Burns |
– |
+ |
– |
+ |
2.024 |
38.41 |
12.6 |
11.91 |
26.875 |
21.84 |
B: burns; EY: eye; W: wound; E: Ear swabs; S: sputum; U: urine
P. aeruginosa virulence is multifactorial and has been accredited to cell-associated factors like alginate, lipopolysaccharide, flagellum, pilus and non-pilus adhesins as well as exoenzymes or secretory virulence factors like protease, elastase, pyocyanin, exotoxin A, exoenzyme S, hemolysins (rhamnolipids and phospholipase) and siderophores.42 Pathogenesis of P. aeruginosa takes place by these factors inducing infections like respiratory tract infections, burn wound infections and keratitis.4 P. aeruginosa produces a number of extracellular components that once colonised can cause significant tissue damage, bloodstream invasion and spread.5 In our study we investigated the relationship between P. aeruginosa QS system and its virulence factors, which have an important role in the pathogenicity of this pathogen.
Pyocyanin synthesis by P. aeruginosa reduces the acute inflammatory response by speeding up neutrophil apoptosis and lowering local inflammation, which is beneficial for bacterial survival.3 In this study, the production of pyocyanin in QS deficient isolates was less than non-QS deficient ones. The Las system regulates the production of LasA, whereas the Rhl system regulates the production of pyocyanin. Pyocyanin production should, however, be regulated by the Las system due to the hierarchical nature of the QS systems and the relevance of the Las system in the hierarchy.29,43 Las and Rhl are two interconnected QS systems that control production of pyocyanin.44,45 A link was discovered between P. aeruginosa QS deficient isolates that were gained from sputum or endotracheal tubes and some virulence factors such as elastase and pyocyanin.46 Moreover, there was another study reported that P. aeruginosa QS deficient strains and strains that produce no or low amount of AIs as N-(3-oxododecanoyl) homoserine lactone and N-butyryl homoserine lactone, produce lower amount of pyocyanin.33
Elastase and protease, which are produced by P. aeruginosa, are extracellular enzymes that aid in colonization of the host.3 Las and Rhl QS systems control the production of alkaline protease and LasA protease in P. aeruginosa.29,43,44 It was observed that major protease and elastase were reduced by deleting the lasR open reading frame of some strains.47 In other studies, P. aeruginosa QS deficient strains were characterized by low protease activity.28,33 Our study has similar outcome, where, P. aeruginosa QS deficient isolates produce lower levels of protease in comparison to the non-QS deficient isolates.
Swimming motility takes place by flagella and type IV pili, where bacteria can incorporate itself in the epithelial cells, and initiate the first step of its colonization on the epithelium.48 Las and Rhl systems in P. aeruginosa take important role in controlling the swimming motility.10,44 Therefore, in our study, the swimming motility of P. aeruginosa QS deficient isolates were lower than that non-QS deficient ones. It is well reported that pyocyanin synthesis, protease production and swimming motility in P. aeruginosa decreased by using febuxostat,12 phenylalanine arginyl b-naphthylamide,11 sodium ascorbate49 and aspirin,50 which all act as QS inhibitors.
Biofilms are formed by adherent cells on cellular or inert substrata and are tangled in 60% of all infections that are characterized by symptoms of moderate severity, chronic development and antimicrobial resistance.51 Previous reports depicted that biofilm formation is controlled by QS.3,52 P. aeruginosa QS is used to initiate growth and produce biofilms.27 A decrease in biofilm formation takes place by sodium ascorbate49, aspirin50 and febuxostat,12 which act as QS inhibitors. Another study indicated that the amount of biofilm formation that was produced by P. aeruginosa QS deficient strains was lower than that of the non-QS deficient strains,33 and this is in accordance with the findings of our study.
Lipase enzyme plays important role in the colonization of P. aeruginosa on human respiratory tract and skin.1,3 While hemolysin, which is a hydrolytic enzyme enhances the spread of bacteria inside the tissues of the host and resistance to the host defence.12 Lipase production is controlled by Rhl system.44 Las and Rhl systems plays important role in lipase and hemolysin production.10 Hemolysins are inhibited by some QS inhibitors as sodium ascorbate,49 febuxostat12 and aspirin.50 According to our study, lipases and hemolysin levels in P. aeruginosa QS deficient isolates were lower than that non-QS deficient ones.
The QS system in P. aeruginosa is important for controlling the production of virulence factors and connected to numerous pathogenic phenotypes including bacterial capacity to attach to tissues and build biofilms. Discovery and development of anti-QS therapeutics are very important to stop or decrease the severity of P. aeruginosa virulence and to overcome the problem of drug resistance; especially P. aeruginosa is one of the most abundant causes of nosocomial infections in developing countries.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Research Ethics Committee of Tanta University, Faculty of Pharmacy, Egypt. Protocol approval code TP/RE /12-21-M-002.
AVAILABILITY OF DATA
All datasets analysed in the study are included in the manuscript and presented as tables and Figures.
- Zhong L, Ravichandran V, Zhang N, et al. Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions. Int J Mol Sci. 2020;21(6):2190.
Crossref - Sutlief AL, Valquier-Flynn H, Wilson C, et al. Live Cell Analysis of Shear Stress on Pseudomonas aeruginosa Using an Automated Higher-Throughput Microfluidic System. J Vis Exp. 2019;(143):58926.
Crossref - Pena RT, Blasco L, Ambroa A, et al. Relationship Between Quorum Sensing and Secretion Systems. Front Microbiol. 2019;10:1100.
Crossref - Babour I, Mohamed M, Shehabi A. Molecular characterization of Pseudomonas aeruginosa isolates from various clinical specimens in Khartoum/Sudan: Antimicrobial resistance and virulence genes. The International Arabic Journal of Antimicrobial Agents. 2020;10.
Crossref - Pang Z, Raudonis R, Glick BR, Lin T-J, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Abdel-Sattar ES, Miyoshi SI, Elgaml A. Regulation of Vibrio mimicus metalloprotease (VMP) production by the quorum-sensing master regulatory protein, LuxR. J Basic Microbiol. 2016;56(10):1051-1058.
Crossref - Elgaml A, Miyoshi S. Role of the Histone-Like Nucleoid Structuring Protein (H-NS) in the Regulation of Virulence Factor Expression and Stress Response in Vibrio vulnificus. Biocontrol Science. 2015;20(4):263-274.
Crossref - Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6(1):26-41.
Crossref - Viducic D, Murakami K, Amoh T, Ono T, Miyake Y. Role of the interplay between quorum sensing regulator VqsR and the Pseudomonas quinolone signal in mediating carbapenem tolerance in Pseudomonas aeruginosa. Res Microbiol. 2017;168(5):450-460.
Crossref - Gala V, Desai KJ. Plant based quorum sensing inhibitors of Pseudomonas aeruginosa. International Journal of Pharmacy. 2014;6(8):20-25.
- El-Shaer S, Shaaban M, Barwa R, Hassan R. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl β-naphthylamide. J Med Microbiol. 2016;65(10):1194-1204.
Crossref - Abbas H, Soliman W, Shaldam M. Perturbation of Quorum Sensing in Pseudomonas aeruginosa by Febuxostat. Adv Microbiol. 2018;08(08):650-664.
Crossref - Elgaml A, Miyoshi S. Presence of Nitric Oxide-Sensing Systems in the Human Pathogen Vibrio vulnificus. Biocontrol Sci. 2015;20(3):199-203.
Crossref - Miyoshi S, Wang J, Katoh K, Senoh M, Mizuno T, Maehara Y. An extracellular serine protease produced by Vibrio vulnificus NCIMB 2137, a metalloprotease-gene negative strain isolated from a diseased eel. World J Microbiol Biotechnol. 2012;28(4):1633-1639.
Crossref - Erickson DL, Endersby R, Kirkham A, et al. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun. 2002;70(4):1783-1790.
Crossref - Storey DG, Ujack EE, Mitchell I, Rabin HR. Positive correlation of algD transcription to lasB and lasA transcription by populations of Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Infect Immun. 1997;65(10):4061-4067.
Crossref - Storey DG, Ujack EE, Rabin HR. Population transcript accumulation of Pseudomonas aeruginosa exotoxin A and elastase in sputa from patients with cystic fibrosis. Infect Immun. 1992;60(11):4687-4694.
Crossref - Storey DG, Ujack EE, Rabin HR, Mitchell I. Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect Immun. 1998;66(6):2521-2528.
Crossref - Wu H, Song Z, Hentzer M, et al. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology (Reading). 2000;146(Pt 10):2481-2493.
Crossref - Geisenberger O, Givskov M, Riedel K, Hoiby N, Tummler B, Eberl L. Production of N-acyl-L-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol Lett. 2000;184(2):273-278.
Crossref - Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 12 2000;407:762-764.
Crossref - Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68(7):4331-4334.
Crossref - Rumbaugh KP, Griswold JA, Hamood AN. Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. J Burn Care Rehabil. 1999;20(1 Pt 1):42-49.
Crossref - Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67(11):5854-5862.
Crossref - Tang HB, DiMango E, Bryan R, et al. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64(1):37-43.
Crossref - Wu H, Song Z, Givskov M, et al. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology (Reading). 2001;147(Pt 5):1105-1113.
Crossref - De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67(4):1865-1873.
Crossref - Zhu H, Bandara R, Conibear TC, et al. Pseudomonas aeruginosa with lasI quorum-sensing deficiency during corneal infection. Invest Ophthalmol Vis Sci. 2004;45(6):1897-1903.
Crossref - Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect 2000;2(14):1721-1731.
Crossref - Collee JG, Miles RS, Wan B. Tests for the identification of bacteria. In J.G. Collee, A.G. Fraser, B.P. Marmion, and A. Simmons (eds.). Mackie and McCartney Practical Medical Microbiology. 14th ed. Churchill Livingstone, Edinburgh ed. 1996:131–150.
- Elgaml A, Hassan R, Barwa R, Shokralla S, Elnaggar W. Analysis of 16S ribosomal RNA gene segments for the diagnosis of Gram negative pathogenic bacteria isolated from urinary tract infections. Afr J Microbiol Res. 2013;7(23):2862-2869.
Crossref - Flanagan TD. A Review of: “Current Protocols in Molecular Biology, Edited by F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, Greene Publishing Associates and Wiley-Interscience John Wiley and Sons, New York. Preparative Biochemistry. 1988;18(3):377-378.
Crossref - Schaber JA, Carty NL, McDonald NA, et al. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004;53(Pt 9):841-853.
Crossref - Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172(2):884-900.
Crossref - MacDonald JC. Pyocyanine. In: Gottlieb D, Shaw PD, eds. Biosynthesis. Springer Berlin Heidelberg; 1967:52-65.
Crossref - Rossignol G, Merieau A, Guerillon J, et al. Involvement of a phospholipase C in the hemolytic activity of a clinical strain of Pseudomonas fluorescens. BMC Microbiol. 2008;8:189.
Crossref - Skindersoe ME, Alhede M, Phipps R, et al. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(10):3648-3663.
Crossref - Kumar S, Kikon K, Upadhyay A, Kanwar SS, Gupta R. Production, purification, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein Expr Purif. 2005;41(1):38-44.
Crossref - Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 25 2000;97(9):4885-4890.
Crossref - Abdi-Ali A, Mohammadi-Mehr M, Agha Alaei Y. Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa. Int J Antimicrob Agents. 2006;27(3):196-200.
Crossref - Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175-179.
Crossref - Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7:212527-212527.
Crossref - de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68(9):4839-4849.
Crossref - Girard G, Bloemberg GV. Central role of quorum sensing in regulating the production of pathogenicity factors in Pseudomonas aeruginosa. Future Microbiol. 2008;3(1):97-106.
Crossref - Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6(1):56-60.
Crossref - Le Berre R, Nguyen S, Nowak E, et al. Quorum-sensing activity and related virulence factor expression in clinically pathogenic isolates of Pseudomonas aeruginosa. Clinical microbiology and infection. Clin Microbiol Infect Dis. 2008;14(4):337-343.
Crossref - Barbieri JT, Van Delden C, Pesci Everett C, Pearson James P, Iglewski Barbara H. Starvation Selection Restores Elastase and Rhamnolipid Production in a Pseudomonas aeruginosaQuorum-Sensing Mutant. Infection and immunity. 1998/09/01 1998;66(9):4499-4502.
Crossref - Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39.
Crossref - El-Mowafy SA, Shaaban MI, Abd El Galil KH. Sodium ascorbate as a quorum sensing inhibitor of Pseudomonas aeruginosa. J Appl Microbiol. 2014;117(5):1388-1399.
Crossref - El-Mowafy SA, Abd El Galil KH, El-Messery SM, Shaaban MI. Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa. Microb Pathog. 2014;74:25-32.
Crossref - Di Pasquale MF, Sotgiu G, Gramegna A, et al. Prevalence and Etiology of Community-acquired Pneumonia in Immunocompromised Patients. Clinical infectious diseases: An official publication of the Infectious Diseases Society of America. 2019;68(9):1482-1493.
Crossref - Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Revi. 2012;76(1):46-65.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.