ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa (P. aeruginosa) is the most important, resistant and dangerous organism in burn wound infections in human. Extracellular and virulence factors lead to tissue damage during infection with P. aeruginosa. One important virulence factor including exotoxin A(ETA) encoded by the tox A gene. In this study , 45 wound swabs from burned skin patients admitted to Al-Hilla Teaching Hospital/Babylon, 2013 from different localities in Babylon were collected to study the frequency of P. aeruginosa and human immune response. Polymerase chain traction technique (PCR) was used for detection ETA gene as a virulent factor producing by P. aeruginosa from burned skin wound infections together with interferon gamma and interleukin 18. The results showed that P. aeruginosa had a frequency of 51.1% among burned skin patients. The bacteriological culture showed that 19 out of 45 (42.2 %) were positive for P. aeruginosa while 23 out of 45 (51.1 %) were positive for P. aeruginosa. Only 17 out of 23 (73.91 %) were ETA producing P. aeruginosa. Also bacteremia occurred only in 9 out of 17 (52.94 %) in ETA producing P. aeruginosa. P. aeruginosa infection was presented to a lesser degree. In addition to that P. aeruginosa that expresses the ETA gene were the most common and toxic pathogens in burned patients and lead to bacteremia and septicemia. The using of PCR technique a rapid and accurate technique might be helpful in combating its toxicity. ETA might contributed to the overall virulent factor.
Bacteremia, Pseudomonas aeruginosa, burn.
Pseudomonas aeruginosa was an opportunistic bacteria able to infect humans in addition to animal1. In human, it could cause severe infections in nearly many tissues. Infection of respiratory tract with P. aeruginosa was a main reason of mortality and morbidity in patients specially those with cystic fibrosis2. In hospitals, P. aeruginosa mostly affected those who were admitted to units of intensive care, burn wound infections and patients with chronic illnesses3. In animals, this pathogen lead to bovine mastitis4. Teat wipes contamination is the major source of infection due to various virulence factors causing its pathogenicity5. P. aeruginosa had ETA, sialidase and exoenzyme S6. This bacteria also had secretory system(a type III) which permits toxins delivery directly into host cell cytoplasm7. ETA was important cause of tissue necrosis since it blocked protein synthesis8. It has been found that interferon gamma is an significant cytokine of protection (innate immunity) of other pathogens like Chlamydia pneumoniae and Toxoplasma gondii but its role in pseudomonas aeruginosa infection is still undiscovered.
In this study, we examined detection of ETA virulence factor gene in P. aeruginosa isolates that isolated from burned skin samples by using Polymerase chain reaction technique that based amplification of ETA gene .
Sample collection
45 wound swabs from burned patients admitted to Al-Hilla Teaching Hospitals /Babylon, 2013 in the sterilized test tubes with aseptic precautions, in agreement with International Dairy Federation standards9 collected from different fields in Babylon Provinces and the samples transferred into Laboratory for further evaluation.
Bacterial isolation
Collected samples were cultured onto blood , nutrient, brain heart infusion broth and MacConkey’s agar10,11. All isolates were gram negative bacteria, with positive oxidase test with positive Api-20 system (bio Mיrieux)12.
Extraction of bacterial genomic DNA
Extraction of Genomic DNA of P. aeruginosa was done using (PrestoTM) ( Geneaid. USA) according to instructions of company. Nano drop spectrophotometer was used to check the results and the extracted materials were stored in refrigerator at -20C until performance of PCR technique.
Polymerase chain reaction
This was done through the use of primers that were specific for ETA gene. These were designed through GenBank (NCBI). Pseudomonas aeruginosa isolate 6 ETA gene, partial cds GenBank: (AF227424.1) through the use of primer3 plus which was online designed. The primers toxA-F primer (CGCAAAGCATCGTCTTCGG) in addition to toxA-R primer (TTCCTGGTCC-TGGGCGTAG), were delivered by (Korea, Bioneer). Then PCR was done according to company instructions.
Interferon gamma and interleukin 18 were assessed using IFN-gamma ELISA kit (BioSource) and Il-18 ELISA kit BioSource, Belgium respectively.
Table (1) showed that the frequency of P. aeruginosa infection was 51.1 % among human burned skin infections. This high occurrence infection rate might be due to the opportunistic nature of this pathogen that lead to its colonization on burned skin when the mechanical or the immunological defect either alone or both were breached14. This was consistent with15 and inconsistent with16 and17 who showed that the frequency of P. aeruginosa was 36%. This might be due to geographical variation.
Table (1):
Frequency of P. aeruoginosa among burned skin infection
Positive P. aeruginosa |
Total |
% |
|
|---|---|---|---|
Human burned skin infection |
23 |
45 |
51.1 |
Because P. aeruginosa was important medically, numerous techniques had been established for quickly recognition of species of P.aeruginosa. Though conventional procedures to recognize P. aeruginosa were dependable, they need numerous days. PCR had the probable chance for microbial species identification quickly through gene sequences amplification specific to P. aeruginosa18.
Table (2) showed that the sensitivity, specificity, PPV and NPV were (80.95 %, 84.61%, 89.47% and 73.3 % respectively). This might be due to true infection by P aeruginosa detected only by PCR(4 cases) with false negative culture results. This might be due to overgrowth of other bacteria or presence of non-cultivable organisms or mutations18,19. Therefore we could conclude that the PCR screening is mandatory technique to detect low number of this organisms at their initial colonization, that is a very important clinically in detection of early infection by this dangerous pathogen when there is no or intermittent colonization with this bacteria.
Table (2):
Sensitivity, Specificity, Positive predictive value and Negative predictive value of bacterial culture
Test |
Sensitivity |
Specificity |
Positive predictive value |
Negative predictive value |
|---|---|---|---|---|
Bacterial culture |
80.95 |
84.61 |
89.47 |
73.3 |
Table (3) showed that 17 out of 23 (73.91%) of human P. aeroginosa produce ETA. As the ETA was very important in P. aeruginosa disease process, so its high frequency in these isolates reflected that it was a prominent virulent factor for this pathogen and our finding was consistent with20,21.
Table (3):
Frequency of ETA among human P.aeruginosa isolated from patients with burned skin
ETA |
Total |
% |
|
|---|---|---|---|
Human P. aeruginosa |
17 |
23 |
73.91 |
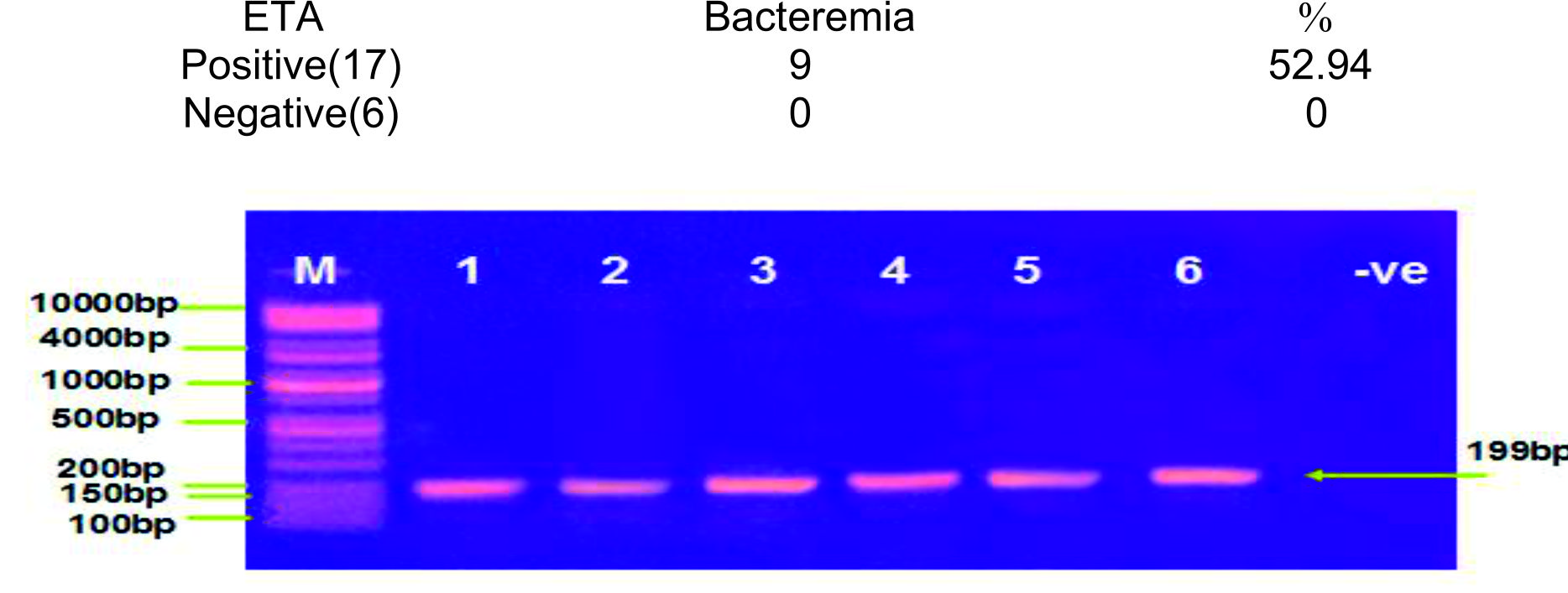
Table (4) showed that 9 out of 17 (52.94%) ETA positive P. aeruginosa had bacteremia while none of the ETA negative P. aeruginosa had bacteremia. This might be due to the major role of ETA inhibition of elongation factor 2 such as what happened with the diphtheria toxin that lead to death of cell22,23 with resultant bacterial dissemination to blood. In addition to that ETA had a cytotoxic effect that extend to varied range of mammalian cells. In addition to that, ETA had the ability of inhibition of macrophage and granulocytes24,25, inhibit the production of IL-1 and altered TNF-alpha production26. All of the above might indicate a major role for ETA in the P. aeruginosa bacteremia and death amongst patients with burned skin. This finding support what was mentioned by other researchers27,28.
Table (4):
showed the frequency of human P. aeruroginosa bactermia in relation to ETA
ETA |
Bacteremia |
% |
|---|---|---|
Positive(17) |
9 |
52.94 |
Negative(6) |
0 |
0 |
Table (5) showed that both interferon gamma and IL-18 was significantly decreased in burned patients with pseudomonas aeruginosa infection in comparison to their value in burned patients without pseudomonas aeruginosa infection (P value < 0.05). IL-18 might act as a stimulus for production of interferon gamma during stimulation by bacterial infection of macrophage to drive Th1 T cell development (33&34). So IL-18 might lead to host resistance through the production of interferon gamma. This was consistent with Xi Huang et al. 201835.
Table (5):
Mean levels of interferon gamma and IL-18 among burned patients in relation to pseudomonas aeruginosa infection
Burned patients with pseudomonas aeruginosa infection |
Burned patients without pseudomonas aeruginosa infection |
P value |
|
|---|---|---|---|
interferon – γ level (pg/ml) |
4.91±2.13 |
11.58±3.11 |
<0.05 |
IL-18(pg/ml) |
33.72±11.66 |
73.94±17.49 |
<0.05 |
- PCR screening is mandatory technique to detect low number of P. aeruginosa infection at their initial colonization, that is a very important clinically in detection of early infection by this dangerous pathogen when there is no or intermittent colonization with this bacteria.
- As the ETA was very important in P. aeruginosa disease process, so its high frequency in these isolates reflected that it was a prominent virulent factor for this pathogen .
- There was a major role for ETA in the P. aeruginosa bacteremia amongst patients with burned skin.
- IL-18 might act as a stimulus for production of interferon gamma during stimulation by bacterial infection of macrophage to drive Th1 T cell development. So IL-18 might lead to host resistance through the production of interferon gamma.
Acknowledgements
None.
Conflicts Of Interest
The authors declare that there is no conflicts of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Engel J.N. Molecular pathogenesis of acute Pseudomonas aeruginosa infections. Severe Infections Caused by Pseudomonas aeruginosa. A.R. Hauser and J. Rello. Dordrecht, Kluwer Academic Publsihers: 2003, pp 201-229.
- Fegan M., Francis P., Hayward A.C., Davis G.H., Furest J.A. Phenotypic conversion of Pseudomonas aeruginosa in cystic fibrosis. J. Clin. Microbiol., 1990; 28: 1143-1146.
- Yetkin G., Otlu B., Cicek A., Kuzucu C., Durmaz R.Clinical, microbiologic, and epidemiologic characteristics of Pseudomonas aeruginosa infections in a university hospital, Malatya, Turkey. Am. J. Infect. Control., 2006; 34: 188-192.
- M. daly 1 E., power 2 j.; bjo¨rkroth3 p., sheehan 2 A., o’connell 2 M., colgan, 2 H.; korkeala,3 and S. fanning1., Molecular Analysis of Pseudomonas aeruginosa: Epidemiological Investigation of Mastitis Outbreaks in Irish Dairy Herds. APPL. ENVIRON. MICROBIOL. 1999; p.; 2723–2729.
- Nikbin V.S.1, Aslani M.M.1, Sharafi Z.1, Hashemipour M.1, Shahcheraghi F.1, Ebrahimipour G.H.2. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran J Microbiol., 2012; 4(3): 118-123.
- Van Delden C., Iglewski B.H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis., 1998; 4: 551-560.
- Rietisch A., Gely V.I., Dove L.S. and Mekalane J.J. “Exs E a secreted regulator of type III sectior genes in Pseudomonas aeruginosa, Journal Harvard Medical School, 2005; 102(22): pp.8006-8011.
- Al-Rubaiee L. “The Role of Pseudomonas aeruginosa in chronic suppurative otitis media infection”. Thesis M.Sc., Medicine collage, University of Baghdad, 2009; pp.5, 6, 20, 22,
- International Dairy Federation: Laboratory methods for use in mastitis work. Brussels: IDF, Document 132, 1981.
- Brooks G.F., Butel J.S., Morse S.A. “Medical Microbiology”. Jawetz,; Melnick and Adelbergs.; 25th ed., McGraw-Hill Companies. Appleton and Lange.; California, 2010.
- Vandepitte J., verhaegen J., Engbaek K., Rohner P., Piot P., Heuck C.C. “Basic laboratory procedures in clinical bacteriology”, 2ed. World Health organization. Geneva, 2003.
- Wolska K. and Szweda P. Genetic features of clinical Pseudomonas aeruginosa strains. Pol. J. Microbiol., 2009; 58(3): 255-60.
- Bauer A.W., Kirby W.M., Sherris J.C., Truck M. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Path., 1966; 45: 493.
- Jawetz E., Melnick J.L., Adelberg E.A., Brooks G.E., Butel J.S., and Ornston L.M. Review of medical microbiology.7thed.Middle East Edition.Appleton and Large Norwak,Connection, 1987.
- Madut N.A., Abdel Gadir A.E., Eljallii I.M., Host determinants of bovine mastitis in semi intensive production system of Khartoum State, Sudan.; J. cell Anim. Biol., 2009; 3(5): 71-77.
- Krik J.H., Bartlett P.C. Nonclinical Pseudomonas aeruginosa mastitis in diary herd. JAVMA, 1984; 184: 671-673.
- Sumathi B.R., Veeregowda B.M., Gomes A.R. Prevalence and antibiogram profile of bacterial isolates from clinical bovine mastitis. Veterinary World, 2008; 1(8): 237-238.
- Xu J., J.E. Moore, P.G. Murphy, B.C. Millar, and J.S. Elborn. ”Early detection of Pseudomonas aeruginosa-comparison of conventional versus molecular (PCR) detection directly from adult patients with cystic fibrosis (CF)”. Annal. Cli. Microbiol. And Antimicro., 2004; 3: pp.21.
- DeVos D., Lim A. Jr., Pirnay J. P., Struelens M.; Vandenvelde C., Duinslaeger L., anderkelen A. and Cornelis P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectoration by multiplex PCR based on two outer membrane lipoprotein genes, opr I and opr L. J.Clin. Microbiol., 1997; 35(6): 1295-99.
- Wolska K. and Szweda P. Genetic features of clinical Pseudomonas aeruginosa strains. Pol. J.Microbiol.,2009; 58(3): 255-60.
- Khan A.A. and Cerniglia C.E. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of exotoxin A gene using PCR, Applied and Environmental Microbiology, 1994; 60: 3739.
- Pavlovskis O.R., Iglewski H., and Pollack M. Mechanism of experimental mouse infections: Adenosine diphosphate ribsylation of elongation factor 2. Infect. Immun., 1978; 19: 29.
- Woods D.E. and Iglewski B.H. Toxins of pseudomonas aeruginosa: New perspectives. Rev. Infect. Dis., 1983; 5: 714.
- Stuart R.K. and Pollack M. Pseudomonas aeruginosa exotoxin A inhibits proliferation of human bone marrow progenitor cells in vitro. Infect. Immun., 1982; 38: 206.
- Pollack M. and Anderson S.E. Toxicity of pseudomonas aeruginosa exotoxin A for human macrophages. Infect.Immun., 1978; 19: 1092.
- Misfeldt M.L., Leggard P.K., Howell S.E., Fornella M.H. and Le Grand R.D. Induction of interleukin-1 from murine peritoneal macrophages by Pseudomonas aeruginosa exotoxin A. Infect. Immun., 1990; 58: 978.
- Gang R.K., Bang R.L., Sanyal S.C., Mokaddas E. and Lari A.R. Pseudomonas aeruginosa septicemia in burns.Burns, 1999; 25: 611.
- Furuya N., Hiralata Y., Tomonok., Matsumoto T., Kaker M. and Yamaguchi K. Rates in mice with endogenous septicemia due to Pseudomonas aeruginosa isolates. J. Med. Microbiol., 1993; 39: 141.
- Greta G., Alvydas P., and Violeta K. The pecularities of Pseudomonas aeruginosa resistance to antibiotics and prevalence of serogroups. Medicina (Kaunas), 2007; 43(1): 36 – 42.
- Shlaes D.M. Sahm D., Opiela C. and Spell berg B. The FDA reboot of antibiotic development. Antimicrobial agents and chemotherapy, 2013; Do1:10.1128/AAC.01277-13.
- Edwin L., Dimatatac M.D., Marissa M., Alejandria M.D., Cecilia Montalban M.D., Cristina Pineda R.M., Concepcion Ang R.M. and Rachel Delino , M.S. (2003). Clinical Outcomes and Costs of Care of Antibiotic Resistant Pseudomonas aeruginosa infections. Phil. J. Microbiol. Infect. Dis., 1999; 32(4): 159 – 167.
- Russell A.D. “Bacterial resistance to disinfectants: present knowledge and future problems”. J. Hosp. Infect., 1999; 43: pp.S57-68.
- Netea M.G., Fantuzzi B.J., Kullberg R.J., Stuyt E.J., Pulido R.C., McIntyre J.L., Joosten J.W., Van D.M. and Dinarello D.C. Neutralization of IL-18 reduces neutrophil tissue accumulation and protects mice against lethal Escherichia coli and Salmonella typhimurium endotoxemia. J. Immunol., 2000; 164: 2644. 18.
- Decken K., G. Kohler K., Palmer-Lehmann A., Wunderlin G., Mattner J., Magram M.K., Gately C. and Alber G. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect. Immun., 1998; 66: 4994.).
- Xi Huang S.A., McClellan R.P., Barrett and Linda D., Hazlett M. IL-18 Contributes to Host Resistance Against infection with Pseudomonas aeruginosa Through Induction of IFN- gamma Production (Journals of immunology,5755-5763), 2018.
- Scharton-Kersten T.M., Wynn T.A., Denkers S., Bala E., Grunvald S., Hieny R.T., Gazzinelli Y. and Sher A. In the absence of endogenous IFN-_, mice develop unimpaired IL-12 responses to Toxoplasma gondii while failing to control acute infection. J. Immunol., 1996; 157: 4045.
- Rottenberg M.E., Gigliotti R.D., Gigliotti M., Ceausu C., Une V., Levitsky N. and Wigzell H. Regulation and role of IFN in the innate resistance to infection with Chlamydia pneumoniae. J. Immunol., 2000; 164: 4812.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.