ISSN: 0973-7510
E-ISSN: 2581-690X
With the ever-increasing pace of urban life, many people now rely a lot on processed foods. Some of these processed foods are prepared hygienically and yet some are prepared in homestead and brought to the open market, without having gone through any microbial quality checks. This has in the recent past led to outbreaks of food -borne diseases such as Cholera and Salmonellosis to mention a few. In this study, processed food samples were collected from three open markets in Windhoek namely Single quarters, Soweto market and the Okuryangava Bus stop. The samples included maize (flour), dried Cleome gynandra leaves, mopane worms, roasted groundnuts, minnos, and cooked beef steaks. Morphological identification was done based on the macroscopic characters based on the colour, shape, and texture and microscopic characterization using lactophenol cotton blue wet mount. DNA was extracted from the fungal isolates and PCR was performed using ITS1 and ITS4 primers. The PCR products were sequenced, and the results were used for identification using the BLAST search program in NCBI. Dried Cleome gynandra contained most of the isolated fungal species which were identified Pichia burtonii, P. macrostoma, Dothidealus, and A. parasiticus. However, Aspergillus species were present in all the food samples and A. parasiticus was found in all food samples except the ground nuts. These could be an indication of that the fungus prefers low water activity and possibly high salt concentrations to survive.
Namibia, Fungal contamination, Processed food, Open markets
Food by its very nature is expected to be nutritious; therefore, food is a rich habitat for microorganisms, in contrast with the great natural systems, soil, water and plants. Given the right physiochemical conditions, only the most fastidious microorganisms are incapable of growth in foods, so that factors other than nutrients usually select for particular types of microbial populations (Pitt and Hockings, 2009). Fungi are well documented organisms that cause food spoilage and have always been a major concern for food experts because of their ability to produce mycotoxins. Fungal contamination makes the food unfit for consumption by causing discolouration, loss of nutrients, heating and mustiness (Bhattacharya and Raha, 2002). The major factors contributing to food contamination are microorganisms, especially fungi which produce volatile compounds, during both primary and secondary metabolism with confirmed properties referred to as mycotoxins. These can be used for detection and identification (Aquino, 2011; Betran and Isakeit 2003). Some fungi cannot be processed out of certain types of food such as xenophiles that can grow in concentrated food e.g Xeromyces bisporus, and some have ascospores that are of very high heat resistance which can survive heat processing e.g Neosartorya fischer (Pitt and Hockings, 2009). The practices of harvesting, handling and production may cause additional contamination and microbial growth. In fact, bacteria and fungi occur naturally as a microflora of fruits and vegetable. Microbial purity is an important aspect of wholesome products, considering that medicinal plants are often collected from wild sources. These products are sold in open-air markets, and most toxigenic molds grow very well in this environment when exposed for long periods in the open markets, without protective packaging, proper temperature maintenance, and moisture control. The presence of toxigenic molds represents a potential risk of mycotoxin contamination. The open markets are popular and used by a large population of Windhoek as they provide staple food at affordable prices than the mainstream retail outlets. Thus, the open markets play a vital role in the survival of the poor people of Windhoek. However, there are certain factors observed in the open markets such as poor personal hygiene, sanitation, and working environment which may contribute to food contamination. These factors result in that the open markets being incapable of protecting the local communities from possible sources of food-borne diseases that occur even in a failure to apply known principles of food safety, for instance basic hygiene and sanitation practices (Abegaz, 2006). Unfortunately, there are not a lot of studies to investigate these issues conducted in Namibia. The importance of this study lies in its ability to help provide empirical data that can be used in a framework development of a strategic plan to improve the safety and the quality of staple food sold on the open markets and to create awareness among the public health of the consumers.
Sampling sites
The samples were collected from three open markets in Windhoek. Maize, ombidi (Cleome gynandra) and Uuhangu (minnose from the Barbus family) were from Okuryangava Bus stop. The cooked beef steak was collected from the Soweto market and Mopane worms and dried hake fish from Single quarters. The samples were taken to the laboratory for further processing.
Isolation and identification of fungi
Ringer solution was prepared, and 9 ml was transferred aseptically into sterilized test tubes. About 1g of each sample collected was ground using a sterile mortar and pestle and was diluted in the Ringer solution. Dilution factors ranged from 10-1 to 10-6. Sabouraud Dextrose agar media were prepared and supplemented with chloramphenicol (500mg/L) to prevent bacterial growth. About 0.1ml of the diluents were inoculated in SDA plates using the spread plate method. For each sample, three replicates were maintained for each dilution factor. The plates were incubated at room temperature and observed daily for growth and sporulation over 7 days. After 7 days of incubation different fungal colonies were transferred onto fresh SDA plates and this was repeated until pure cultures were obtained and then a single spore culture was made from the pure culture.
The fungi isolated were characterized based on the macroscopic (color, texture, and shape) and microscopic features. The microscopic identification was done using Lacto phenol cotton Blue (LCB). A drop of 70% ethanol was placed on a clean glass slide and a loopful of the fungal colony was taken and placed on the slide and spread evenly in the ethanol with the help of a sterile needle. Than two drops of LCB was placed onto the slide and the cover slip was gently placed over the drops of LCB. The slide is then observed under 1000x magnification and identified based on morphology characteristics.
DNA extraction
DNA extraction was done using the ZR fungal/bacterial DNA extraction kit that was obtained from the Inqaba Biotechnological Industries (South Africa). However, few modifications were made to the protocol. The modifications are as indicated. Approximately 1g of the fungi was added into the ZR Bashing BeadTM Lysis Tube and 750µl of Lysis solution added to tube. The tubes containing the mixtures were then sonicated at a temperature of 36ºC for 5min to aid in lysis of cells as well as to dislodge the fungal cells from each other. After sonication, the solution was then vortexed for 15min at 2000rpm to allow the optimal lysis of the fungal cells in the sample. The ZR Bashing BeadTM Lysis Tube was then centrifuged at 10000rpm in a micro centrifuge for 1 min and 400µl of the supernatant was transferred to a Zymo-SpinTM IV Spin Filter in a collection tube. The tube was then centrifuged at 7000rpm for 1min.
To the filtrate in the collection tube, 1200µl of Soil DNA Binding Buffer was added and then 800µl of the mixture was transferred to the Zymo-SpinTM IIC Column in a collection tube and centrifuged at 7000rpm for 1min. The flow through was then discarded and the step repeated. 200µl of DNA Pre-Wash Buffer was added to the Zymo-SpinTM IIC Column in a new collection tube and centrifuged at 10000rpm for 1min. Then 500µl of DNA Wash Buffer was added to the Zymo-SpinTM IIC Column in a collection tube and centrifuged at 10000rpm for 1min. The Zymo-SpinTM IIC Column was transferred to a sterile 1.5ml ependorf tube and 100µl of DNA Elution Buffer was added directly to the column matrix and centrifuged at10000rpm for 30sec to elude the DNA. The DNA was collected in the ependorf tube and stored at 4ºC.
PCR Amplification and Sequence Analysis
PCR was performed on DNA obtained. Amplification was performed in a 50-µl reaction mixture containing 100 µM deoxynucleoside triphosphates, 0.1 µM each primer, 1×PCR buffer with 2.0 mM MgCl2, 2 µl of undiluted fungal genomic DNA sample, and 1 unit of Taq polymerase (Qiagen) and 22.5µl of nuclease free water. The thermal cycling program was as follows: The first cycle of initial denaturation was performed at 95°C for 10 min, followed by 30 cycles in series of 95°C for 1 min, 55°C for 1min, and 72°C for 90 s, with a final cycle at 72°C for 10 min (Kumar and Shukla, 2005). The PCR products were sent to the Inqaba Biotechnological Industries (South Africa) for sequencing. Then the sequences similarity searches were performed for each fungal sequence against the nucleotide collection (nr/nt) database maintained by the National Center for Biotechnology Information using the BLASTn algorithm.
Identification of fungi
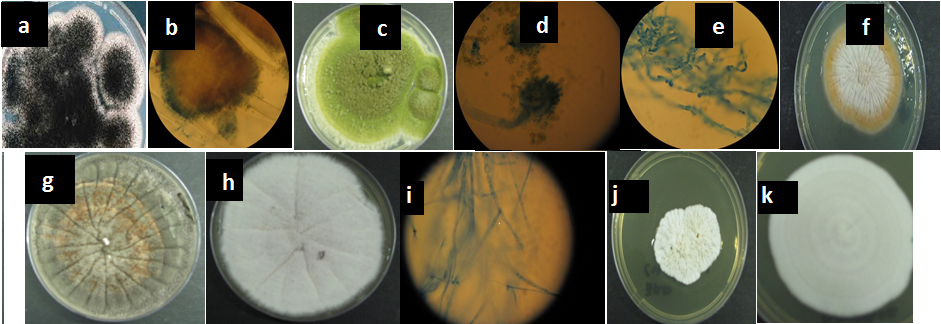
There were 17 different fungal isolates from the food samples. The fungal isolates were first identified based on their colony morphology (Figure 1) and microscopic observations. The species identified were Pichia burtoni, Phoma macrostoma, Dothidealus, Dothideomycetes, Aspergillus parasiticus, Cladosporium tenuissima, Aspergillus niger, Fusarium oxysporum, Rhizopus delemar, Penicillium corylophylum, Fusarium biseptatum, Trichosporon insectorum, Aspergillus flavum, and Trichosporum asahii. The most frequent species were Aspergillus niger and Aspergillus parasiticus. The details of each morphologically and molecularly identified taxon were given below.
Fig. 1. Cultured fungal species from various processed food. The identity of the fungil are represented by letters a to k: a= Aspergillus niger on SDA media; b= Aspergillus niger conidia; c= pure culture of Aspergillus flavus; d= Conidia of Aspergillus parasiticus; e= pure culture of Fusarium biseptatum on SDA media, f= A-pure culture of F biseptatum on SDA media, g= pure cultures of Fusarium oxysporum on SDA media; h= pure cultures of Fusarium ox-ysporum; i= pure culture of Penicillium corylophilum on SDA media; j= Trichosporon. insectorum , k= Trichosporon asahii
For Aspergillus flavus, the colonies start being white to pale yellow, but they quickly form black conidia yet for A. parasiticus, the colonies are wooly and green. The conidiophores are long, rough beneath the globose vesicle. For Fusarium oxysporum, the colonies start of as white but then become tinge with pick and lavender but the lavender reverses to purple yet for Trichosporon colonies are yeast-like, rapid growing, smooth, wrinkled, raised, folded, glabrous to velvety, dull, brittle, waxy, white, or yellowish white to cream colored. The wrinkled appearance becomes more prominent in time. Heaping at the center of the colony is typical. The colonies of Phoma grew rapidly. They are flat, spreading, powdery to velvety, and often largely submerged in the medium. From the front, the color was initially white and later becomes olive grey with an occasional tint of pink. From the reverse, it had dark brown to black. Cladosporium fungi have the following typical characteristics: Conidiophores tall, dark, upright, branched variously near the apex, clustered or single; conidia (blastospores) dark, 1- or 2-celled, variable in shape and size, ovoid to cylindrical and irregular, some typically lemon-shaped; often in simple or branched acropetalous chains (Hawksworth, 1991; Hawksworth 2001, Eva et al., 2010).
Table (1):
Genbank accession numbers and their top BLAST match sequences of the fungal isolates.
Fungal Identity |
GenBank accession number |
Query coverage (%) |
Maximum sequence identity (%) |
|---|---|---|---|
Pichia burtoni |
EU714323.1 |
100 |
100 |
Pleosporalus sp. |
HE820773.1 |
100 |
99 |
Phoma macrostoma |
DQ474112.1 |
100 |
99 |
Dothidealus sp. |
JQ761375.1 |
100 |
100 |
Dothideomycetes sp. |
EU680549.1 |
100 |
100 |
Phoma macrostoma |
DQ474111.1 |
100 |
100 |
Phoma macrostoma |
GQ352490.1 |
100 |
100 |
Aspergillus parasiticus |
JQ316518.1 |
100 |
100 |
Cladosporum tenuissimum |
JX310566.1 |
100 |
100 |
Aspergillus niger |
JX556221.1 |
100 |
100 |
Rhizopus delemar |
JN315024.1 |
100 |
100 |
Penicillium corylophilum |
HQ829133.1 |
100 |
100 |
Fusarium biseptatum |
EU926251.1 |
100 |
92 |
Trichosporon insectorum |
JF825461.1 |
100 |
100 |
Aspergillus flavus |
JX231008.1 |
100 |
100 |
Trichosporon insectorum. |
JF781466.1 |
100 |
100 |
Trichosporon asahii |
JQ863250.1 |
100 |
100 |
PCR and Sequence Analysis
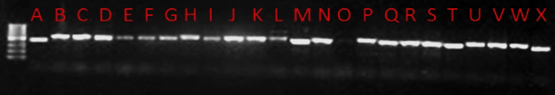
The PCR amplicons (see figure 2) were quantified with 1% agarose gel electrophoresis. Amplification of DNA samples with ITS1-ITS4 primers resulted in fragments of approximately 550bp.
Fig. 2. LANE A, P. burtoni; lane B, Pleosporalus sp.; lane C, P. macrosto-ma; lane D, Dothidealus; lane E, Dothideomyces; lane F, P. macrosto-ma; lane G, P. macrostoma; lane H, A. parasiticus; lane I, C. uredinico-la; lane J, A. parasiticus; lane K, A. niger; lane M, F. oxysporum; lane N, A. parasiticus; lane O, no DNA; Lane P, Rhizopusdelemar, Lane Q, P. corylophilum; Lane R, A. parasiticus; Lane S, Fusarium biseptatum; Lane T, Trichosporoninsectorum; Lane U, A. niger; Lane V, A. niger; Lane W, A. flavus, Lane X, Trichosporon sp
The aim of this study was to establish the identity fungi contamination the ever-increasing presence of processed foods in open markets and the consequently comment on the safety status of these processed foods. A wide range of processed foods where examined in this study and the study revealed the contamination of processed food from the open markets of Windhoek by different fungi, namely: A. flavus, A. niger, A, parasiticus, C. tenuissimum, Dothideomyces, F. oxysporum, F. biseptatum, P. burtonii, P. corylophilum, Pleosporalus sp., Rhizopus delemar, Trichosporon insectorum, and T. asahii. The most common fungi in the food tested were A. niger, A. parasiticus, and A. flavus although A. parasiticus was not found in groundnuts and the cooked beef steak which confirm that this fungus prefers low water activity and higher salt concentrations in order to grow. A. flavus and A. niger were both found in mopane worms but A. flavus was absent in groundnuts. On the other hand, Penicillium corylophilum and Rhizopus delemar were only present in minnose sample and T. asahii and T. insectorum were only present in mopane worms and Cladosporium tenuissimum was only found in the beef steak. The most common and prevalent species in Cleome gynandra were P. burtonii, Phoma macrostoma, Dothidealus and Dothideomyces and Pleosporalus. The F. oxysporum was also present in groundnuts but F. biseptatum was the only species found in nongongo (Manketii nuts). In general, after processing, these food samples are packed in unsterilized containers and transported to the open markets for trading purposes. Moreover, in the open markets, unpackaged foods are displayed in open basins or bowls for sale. With the surrounding environment usually hot and humid, these practices are potential sources of fungal contamination, which may predispose the product to public health hazards (Perrone et al., 2007; Bhat and Vasanthi, 2003; Lewis et 2015; Muriuki and Siboe, 1995).
Some fungi are soil-borne because of their abundance in the soil and their frequent association with plant roots, as either parasites or saprophytes. They can also be air-borne, because they have many active or passive means of dispersal in the atmosphere and are the common colonizers of aerial plant parts. The widespread distribution of these fungi may be attributed to their ability to grow on a wide range of substrates and their efficient mechanisms for dispersal (Samson et al., 2004). This versatility of these fungi has implications of their ability to spread in the human population if they get in contact with them. Groundnuts are always in contact with soil populations and seeds are frequently invaded by soil fungi before harvest (Horn and Greene, 1995). Infection by A. niger and A, flavus occurs under both preharvest and post-harvest conditions. During preharvest conditions infection by A. flavus and A. parasiticus results in consequent aflatoxin contamination being a major problem in semi- arid and tropical environments (Sekar et ., 2008; Shepard, 2008; Wagacha and Muthomi, 2008; Rajeev et al, 2008; Levetin and McMahon, 2003).
PCR was used to amplify the ribosomal DNA, using the universal primers, internal spacer ITS 1 and ITS 4, and these confirmed the presence of the species that were identified. However, other unculturable fungi that may have been potentially present could not be detected. The results also confirmed the genetic variation between the different species was minimal with a common banding pattern that ranged from 490 to 570 bp (see figure 2). The ribosomal RNA genes possess characteristics that are suitable for the detection of pathogens at the species level. These rDNA are highly stable and exhibit a mosaic of concerned and diverse regions within the genome (Manish and Shukla, 2005, White et al.,1990; Rainiere et al., 2003, Vilgalys and Gonzalez, 1990)
In conclusion, processed food samples from the open markets available to consumers were contaminated with known aflatoxigenic forms of Aspergillus species as well as Penicillium, Cladosporium, Trichosporon, Fusarium, Rhizopus delemar, Phoma, Dothideomyces and Pichia burtonii species. This report is the first to be done in Namibia and already has important implications on food safety regulations. Furthermore, from this study it is recommended that research to determine faster and more reliable methods to detect fungal contaminants and be able to develop ways. If precautions are not taken during preharvest and post-harvest of these foods from the buying and packaging to the selling point in the open markets, fungal contamination could raise serious concerns related to environmental, safety, food quality and human health.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Abegaz M. “Food Safety in Food Security and Nutrition, its Impact in Millennium Development Goals Status, Challenges & the Way forward in Developing Countries” Including Food safety and HIV/AIDS. Inaugural Conference of Food & Nutrition Society of Ethiopia (FoNSE), 24-26 January 2006, Addis Ababa, Ethiopia 2006.
- Aquino S. Gamma radiation against toxigenic fungi in food, medicinal and aromatic herbs. In: Science against microbial pathogens: communicating current research and technological advances (A. Méndez-Vilas (Ed.) 2011.
- Betran, F. J., and Isakeit, T. Aflatoxin Accumulation in Maize hybrids of different maturities. Agronomy Journal, 2003; 96: 565-570.
- Bhattacharya K, Raha S. Deteriorative changes of maize, groundnut and soybean seeds by fungi in storage. Mycopathol. 155:135-141. Bryan GT, Daniels MJ, Osbourn AE (1995). Comparison of fungi within the Gaeumannomyces-phialophora complex by analysis of ribosomal DNA sequence. Appl. Environ. Microbiol., 2002; 61: 681-689.
- Bhat, R. V. And Vasanthi, S. Food Safety and Food Security and Food Trade –Mycotxin food safety risk in developing countries. International Food Policy Research Institute 2003.
- Eva Bellemain, T. Carlsen, C. Brochmann, E. Coissac, P. Taberlet, and H. Kauserud. ITS as an environmental DNA barcode for fungi, an in silico approach reveals potential PCR biases. BMC Microbiology, 2010: 10-189.
- Hawksworth D.L. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycology Research, 1991; 105: 1422-1432
- Hawksworth D.L. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycology Research, 2001; 95: 645-655.
- Horn B. W., and R. L. Greene, Vegetative compatibility within the population of Aspergillus flavus, A. parasiticus and A. tamari from peanut field. Mycologia, 1995; 85: 324-332
- Levetin E and McMahon K 2003. Fungi and Human Health: Drugs, Poison, Pathogens, and Allergies (3rd Edition).
- Lewis, L., Onsongo, M., Njapau, H., Schurz-Rogers, H., luber, G., Nyamongo, S. J., Baker, L., Dahiye, A. M, Misore, A, Kevin, D. R, and the Kenya aflatoxin investigating group. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in Eastern and Central Kenya. Environmental Health Perspective, 2005; 113(12): 1763-1767.
- Manish K and Shukla PK. Use of PCR targeting of internal transcribed spacer regions and single-stranded conformation polymorphism analysis of sequence variation in different regions of rRNA genes in fungi for rapid diagnosis of mycotic keratitis. Fermentation Technology Division, Central Drug Research Institute, Lucknow, India. Journal of Clinical Microbiology, 2005: p. 662-668.
- Muriuki, G. K., and Siboe, G. M. Maize flour contaminated with toxigenic fungi and mycotoxins in Kenya. African Journal of Health Sciences, 1995; 2: 236– 241
- Perrone G., Susca A., Cozzi G., Ehrlich K., Varga J., Frisvad J.C., Meijer M., Noonim P., Mahakarnchanakul W., Samson R.A. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol., 2007; 59(1): 53-66.
- Pitt J. I. and Ailsa D. Hockings. Fungi and Food Spoilage 2009.
- Rajeev Bhat, Ravishankar V. Rai, and A.A. Karim. Mycotoxins of Food and Feed: Present status and Future concerns. In: Comprehensive Reviews In Food Science And Food Safety, 2010; 9: pages 57-81.
- Rainiere S, Zambonelli C. and Kaneko Y. Saccharomyces sensu stricto: systematic, genetic diversity and evolution. Journal of Bioscience and Bioengineering, 2003; 96: 1-9. Cambridge UK
- Samson R.A., Hoekstra E. S., Frisvad J. C. Introduction to food and air borne fungi. 7th edition. Centraal Bureau Voor Schimmel cultures Publisher- Utrecht Netherlands 2004.
- Sekar P., N. Yumnam and K. Ponmurugan, K. S. R. 2008. College of Arts and Science, Tiruchengode, Tamil Nadu, India: Screening and Characterization of Mycotoxin Producing fungi from dried fruits and grains. 2008, Advanced Biotechnology, page 12-15.
- Shephard, G. S. Impact of mycotoxins on human health in developing countries. Food Additives and Contaminantsm, 2008; 25(2): 146 – 151.
- Vilgalys R., Gonzalez D. Organization of ribosomal DNA in the basidiomycete Thanatephorus praticola. Current Genetics, 1990; 18: 277-280.
- Wagacha, J. M. and Muthomi, J. W. Mycotoxin problem in Africa: Current Status, implications to food safety and health and possible management strategies. International Journal of Food Microbiology, 2008; 124: 1-12.
- White, T. J., T. Bruns, S. Lee, and J. Taylor. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322. In M. A. Innis, D. H. Gelfand, J. J., Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, California 1990.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.