ISSN: 0973-7510
E-ISSN: 2581-690X
Bio-deteriorated waste is the leftover organic matter of unwanted raw food which if not handled properly or left for natural degradation can cause health issues. Microorganisms have the ability to biodegrade waste by secreting enzymes. The aim of the work was to isolate and identify lipase-producing bacteria from waste polluted (Bio-deteriorated waste and Municipal Solid Waste) dumping sites of Gandhinagar, Gujarat, India. Lipase-producing bacteria were isolated using tributyrin agar as a selective medium. Out of 7 bacterial isolates, 1 isolate (HAL-2) gave the highest lipolytic activity. HAL-2 was identified as Bacillus pumilus by 16S rRNA sequencing. The bacterial isolate gave maximum lipase activity (0.68 U/mL) at 37°C and pH 7.0 Culture medium parameters such as carbon source, nitrogen source, pH, and inoculum size were varied for the purpose of optimization. The maximum lipase production was observed at pH 7.0, 37°C temperature. Inoculum size had an effect of direct proportionality on lipase activity. Glycerol tributyrate was found to be the best substrate (0.68 U/mL). Sucrose and Tryptone in the medium increased enzyme production when compared with other carbon and nitrogen sources.
Bio-deteriorated Waste, Screening, Lipase Producing Bacteria, 16S rRNA Gene Sequencing, Optimization Study
Developed countries create 521.95–759.2 kg of waste per person per year (kpc), compared to 109.5–525.6 kpc in developing countries. According to the most recent projections, global MSW (Municipal Solid Waste) output is approaching 2 billion tonnes per year, creating an environmental hazard1,2.
Every day, Gujarat state, India, produces 8,336 metric tonnes of urban solid trash, while national total solid waste generation is 1,33,760 metric tons per day in urban areas. Gandhinagar, known as the Green city of the Gujarat state, generate approximately 60 tons per day (TPD) Municipal Solid Waste (MSW)3.
Enzymes are used as biological catalysts without any environmental impact because of their high specificity and economic advantages. Enzymes are highly specific. Enzymes can be immobilised and reused multiple times. Enzymes can also be used to remediate trash that contains hazardous substances. Enzymes naturally disintegrate with the aid of decomposers, allowing the enzymes’ whole chemical components to be returned back into nature4. Enzymes are biomolecules that catalyzes, which means, increase the rates of chemical reactions. Most enzymes are proteins. Nearly all metabolic processes in the cell need enzyme catalysis in order to carry out chemical reactions5-7.
Because of their catalytic activity, large yields, and ease of genetic modification, microbial enzymes are more valuable than enzymes originating from plants or mammals.8 Some enzymes are used commercially in Dairy Industries, Pharmaceuticals Industries, Textile Industries, Brewing Industries, synthesis of antibiotics, etc.9,10
Lipase (E.C 3.1.1.3) is known as triacylglycerol acyl hydrolase and belongs to the hydrolases family. In an oil-water interface, lipase catalyses the hydrolysis of triglycerides, turning them to glycerol and fatty acids.11,12 Food, dairy, flavour, medicines, biofuels, leather, cosmetics, detergents, and chemical manufacturers industries all utilise lipases. Lipases can be obtained from plants, animals, and microbes, however, microbial lipases are the most commonly employed class of enzymes in biotechnological and organic chemistry.9,13,14
Sample Collection and Physicochemical Analysis
Soil samples were collected from the Gandhinagar Municipal Corporation (GMC) dumping site from the depth of 10 cm. Bio-deteriorated waste samples were also collected from the vegetable market garbage, sector-21 of Gandhinagar. 50 gm of each selected sample was collected in sterile polythene bags and brought to the laboratory and kept at 4°C for further processing and use.
The physicochemical parameter Temperature, pH, Moisture content, Total carbon content and Total nitrogen content was analysed by standard methods.3,13
Screening and Identification of Bacterial Isolate
1 gm of soil and bio-deteriorated waste samples were suspended in 9.0 mL sterile distilled water. The sample were serially diluted up to 10-7 dilution. 0.1 mL of each dilution was plated on Tributyrin agar plates containing 0.5% (w/v) peptone, 0.3% (w/v) yeast extract, 1% (v/v) Tributyrin and 1.5% agar, pH 7.0) by spread plate method15. Microbial colonies, which appeared on Tributyrin agar plates were further sub cultured on the respective media. The pure Isolates obtained were transferred on Tributyrin agar slants and preserved at 4-5 °C.
Identification of Lipase Producing Bacteria
The potent bacterial isolate showing maximum zone of clearance was selected for further analysis.16.17 Morphological characteristics of the bacterial isolate were studied for the identification of the isolate.18,19 For molecular characterization the bacterial isolate was submitted to Gujarat Bio-technology Research Centre (GBRC), Gandhinagar for Partial 16S rRNA gene sequencing.
Lipase Activity Test
Lipase activity was carried out by the titrimetric method. Glyceryl tributyrate present in Tributyrin broth was used as a substrate. 1 mL culture was added to 100 mL Tributyrin broth and incubated for 24 hrs at 37 °C. After incubation 15 mL mixture was centrifuged (REMI) at 5000rpm for 15 minutes. To 10 mL supernatant 2-3 drops of phenolphthalein indicator were added and then titrated against 0.01N NaOH.20-23
Optimization of Media Parameters for High Enzyme Activity
Effect of Temperature on Lipolytic Activity
For selection of optimum temperature for the production of lipase the temperatures varying from 25 °C-48 °C were selected by maintaining the other parameters constant.24,25
Effect of pH on Lipolytic Activity
To study the effect of pH, the lipase activity was measured at various pH ranging from 6.0 to 8.0 while the other parameters remained unchangesd.24
Effect of Inoculum Size on Lipolytic Activity
1 % to 5 % inoculum was inoculated in Tributyrin broth to study the effect of inoculum size. Bacterial cultures were incubated at 37°C in an orbital shaker with a 100-rpm agitation speed. After 6 hours, the culture broth was collected by centrifugation at 8000xg for 20 minutes at 4°C. The supernatant was tested for enzyme activity and utilized as a crude enzyme solution.24
Effect of Carbon Sources as Lipolytic Activity
To study the effect of carbon sources on lipase production different carbon sources such as sucrose, fructose, mannitol, glucose, and starch were used. The carbon source present in the production media was substituted at the concentration of 1.0 %.24
Effect of Nitrogen Sources as Lipolytic Activity
The effect of nitrogen source was studied by using different nitrogen sources like tryptone, ammonium sulphate, ammonium chloride, ammonium nitrate and potassium nitrate. The nitrogen present in the production media was replaced at the concentration of 1.0 %.24
Effect of Substrate on Lipolytic Activity
Glycerol tributyrate, groundnut oil, sunflower oil, soybean oil, and cottonseed oil were added to the production medium at the concentration of 1.0% to study the effect of different substrates on lipase activity.24
Effect of Metal in on Lipolytic Activity
CuSO4, MgSO4, Zinc, KCL, FeSO4 were added to the production medium at a final volume of 1 mM to study the effect of different metal ions on lipase activity.
Physicochemical Test of Samples
The temperature of the soil sample was 35.2°C in the Pirana site and 35.5 °C in the Gyaspur site1. Whereas in our study the temperature of the soil samples was 38°C, 32°C, 40°C, and 39°C of samples A1, A2, A2, and A4, respectively, and bio-deteriorated waste sample’s temperature was 32°C. The pH of GMC dumping site soil sample A1, A2, A2, and A4 was 6.73, 6.58, 6.51, and 6.07 respectively and bio-deteriorated waste sample B was 6.16 which is almost same as that obtained by K.R. Atalia and his colleagues which was 6.5 and 6.6 respectively. The moisture content of the sample was 25.80 % and 28.40 % in Pirana site and Gyaspur site respectively in study of K.R Atalia et al3 but in our study moisture content of the soil samples was 11.95 %, 8.78 %, 18.98 %, and 18.60 % of samples A1, A2, A2, and A4 respectively and moisture content of the bio-deteriorated waste sample was 42.60 %. In our study C/N ratio of soil samples of GMC dumping site was 1.50 %, 1.32 %, 1.45 %, and 1.64 of samples A1, A2, A2, and A4 respectively and C/N ratio of bio-deteriorated waste sample of vegetable market garbage site was 0.89 % (Table).
Table:
Physicochemical properties of selected samples.
Sample name |
Temp. |
pH |
Moisture Content |
Total Carbon Content |
Total Nitrogen Content |
C/N Ratio |
|---|---|---|---|---|---|---|
A1 |
38 °C |
6.73 |
11.95 % |
0.72 % |
0.48 % |
1.50 % |
A2 |
37 °C |
6.58 |
8.78 % |
0.65 % |
0.49 % |
1.32 % |
A3 |
40 °C |
6.51 |
18.98 % |
0.90 % |
0.62 % |
1.45 % |
A4 |
39 °C |
6.07 |
18.60 % |
0.97 % |
0.59 % |
1.64 % |
B |
32 °C |
6.16 |
42.60 % |
1.71 % |
1.92 % |
0.89 % |
Screening and Identification of Bacterial Isolate
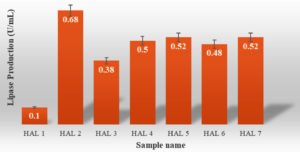
A total of 7 morphologically different bacterial strains were isolated from GMC dumping site and Bio-deteriorated waste samples collected from Vegetable market garbage sites, Gandhinagar, Gujarat, India. The isolated strains were screened for lipase production using Tributyrin agar media. These isolates were identified based on cultural and morphological characterization. They were named as HAL 1, HAL 2, HAL 3, HAL 4, HAL 5, HAL 6, and HAL 7. Out of the 7 bacterial isolate, HAL 2 was found to be potent as it produced larger clear zone (23 mm) than the others, indicating higher lipase activity. The colony was medium in small and irregular in shape and margin it was convex and moist and opacity was opaque and pigmentation was off white. The bacterial isolate HAL 2 was Gram positive and big rod in shape.
The selected bacterial isolate HAL2 was confirmed by sequencing of 16S rRNA gene identified as Bacillus pumilus by Gujarat Biotechnology Research Centre, Gandhinagar, Gujarat. The 16S rRNA gene was amplified and the resultant gene product was found to be 863 BP. The 16S rRNA gene PCR product was sequenced, and the results were deposited in GenBank under the accession number MT539145.
Lipase Activity Test
Out of the 7 bacterial isolate, HAL 2 bacterial isolate gave maximum lipase activity (0.68 U/mL) at 37°C and pH 7.0 (Figure 1) and it was selected for further studies. The lipase activity was low as compared to studies done by R. Sangeetha et al.24
This may be because lipase production is influenced by the type and concentration of carbon and nitrogen sources, the culture pH, the growth temperature and dissolved oxygen concentration20,26 Lipidic carbon sources appear to be required for high lipase yields in most cases; nevertheless, a few researchers have reported good yields in the absence of fats and oils.22
Optimization of Media Parameters for High Enzyme Activity
Effect of Temperature on Lipolytic Activity
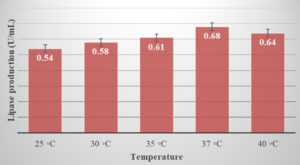
In the present study 37°C was found to be the optimum temperature for lipase activity (0.68 U/mL) Figure 2. The results show that lipase activity increases up to 37°C and then declines above the optimum temperature. Because most organisms’ enzymes rapidly denature at severe temperatures, the decrease in lipase productivity might be attributed to insufficient temperature for microorganism growth and enzyme production. Similar results were reported in the study of R. Sangeetha et al24 and Sagar et al25 that the maximum lipase production was at 37°C.
Effect of pH on Lipolytic Activity
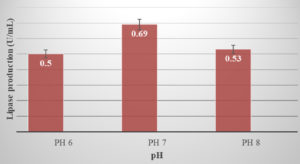
The maximum lipase activity (0.69 U/mL) was observed at pH 7.0, (Figure 3). R. Sangeetha et al13, reported that the lipase activity for Bacillus pumilus was highest at pH 9.0. Similarly, Lipase produced by Bacillus pumilus SG2 exhibited maximum activity at alkaline pH. According to R. Sangeetha et al24, the enzymes were stable between pH 7.0 and 11.0.
Effect of Inoculum Size on Lipolytic Activity
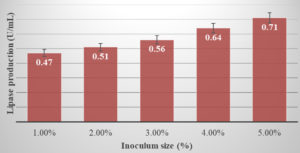
In our study, it was observed that there is a direct relationship with inoculum size and lipase activity. There was an increase in the lipase activity as the inoculum size increased. (Figure 4).
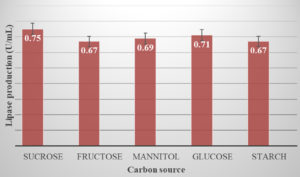
Effect of Carbon Sources as Lipolytic Activity
The carbon source present in the media plays an important role in lipase activity. The lipase activity was found to be maximum for sucrose (0.75 U/mL) followed by glucose (0.71 U/mL) and mannitol (0.69 U/ml) while optimizing the process for carbon source. The different carbon sources were added to the production medium at the concentration of 1%. (Figure 5). Similar results were obtained by R. Sangeetha et al24 in which Sucrose was found to be an effective carbon source for the lipase production.
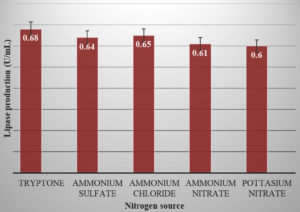
Effect of Nitrogen Sources as Lipolytic Activity
Beside carbon source, the type of nitrogen source in the medium also influences the lipase activity in production broth. In our study, the lipase activity was found to be maximum for tryptone (0.68 U/ml) (Figure 6). Microorganisms mostly show high yield of lipase activity when organic nitrogen sources are used. The inorganic nitrogen sources i.e. Ammonium chloride also showed its influence on lipase activity. R. Sangeetha et al24 in their study found Casein and yeast extract to be effective in lipase production.
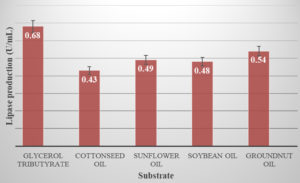
Effect of Substrate on Lipolytic Activity
HAL-2 showed maximum lipase activity when the media was supplemented with 1% Glycerol tributyrate. Apart from these, groundnut oil, sunflower oil, soybean oil, and cottonseed oil also showed positive effect on lipase activity but it was less in comparison to Glycerol tributyrate. Glycerol tributyrate can also be considered as a best carbon source for lipase production. (Figure.7)
R. Sangeetha et al11, in their study found olive oil, coconut oil and castor oil to be the best crude substrates for lipase production. According to Ertuğrul S. et al27 Olive oil was the best carbon source for Bacillus sp. for lipase production.
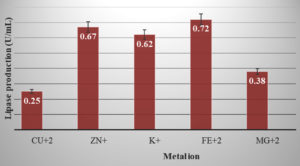
Effect of Metal ion Lipolytic Activity
HAL2 showed maximum lipase activity in the presence of Fe2+ ion in our study. Zn+ and K+ also showed significant effect but it was less in comparison Fe2+. Mg2+ and Cu2+ showed the least effect (Figure 8).
From the present study lipase producing bacteria were isolated from the dumping waste site and bio-deteriorated waste. Out of the total 7 isolates obtained bacterial isolate HAL 2 exhibited the highest lipase activity and was characterized as Bacillus pumilus S113 through 16s rRNA partial sequencing. Lipase activity was determined by titrimetric method with higher activity of 0.67 U/mL at 37°C at pH 7.0. Optimization of culture conditions such as pH, temperature, inoculum size and various substrate were studied for production of lipase. 1 % sucrose and tryptone were used as carbon and nitrogen source respectively. Limitations of the industrial use of these enzymes have mainly been due to their high production costs, which may be overcome by molecular technologies.
ACKNOWLEDGMENTS
The authors would like to thank Gandhinagar Municipal Corporation (GMC) for giving us permission to collect the required necessary sample for research work. We also thank Gujarat Biotechnology Research Centre (GBRC), Gandhinagar for the identification and 16s rRNA Sequencing.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analysed during this study are included in the manuscript.
ETHICS STATEMENT
The article does not contain any studies with human participants or animals by any of the authors.
- Karak T, Bhagat RM, Bhattacharyya P. Municipal solid waste generation, composition, and management: The world scenario. Crit Rev Environ Sci Technol. 2012;42(15):1509-1630.
Crossref - Nandan A, Yadav BP, Baksi S, Bose D. Recent Scenario of Solid Waste Management in India. World Sci News. 2017;66:56-74. www.worldscientificnews.com.
- Atalia KR, Buha DM, Joshi JJ, Shah NK. Microbial biodiversity of municipal solid waste of ahmedabad. J Mater Environ Sci. 2015;6(7):1914-1923.
- Berg, J. M., Tymoczko J. L. LS. Biochemistry 5th Edition. W.H Freeman and Company; 2022.
- Seitz EW. Industrial application of microbial lipases: A review. J Am Oil Chem Soc. 1974;51(2):12-16.
Crossref - Eltaweel MA, Rahman RNZRA, Salleh AB, Basri M. An organic solvent-stable lipase from Bacillus sp. strain 42. Ann Microbiol. 2005;55(3):187-192.
- Jaeger KE, Eggert T. Enantioselective biocatalysis optimized by directed evolution. Curr Opin Biotechnol. 2004;15(4):305-313.
Crossref - Marimuthu K. Isolation and characterization of Staphylococcus hominis JX961712 from oil contaminated soil. J Pharm Res. 2013;7(3):252-256.
Crossref - Celligoi MAPC, Baldo C, De Melo MR, Gasparin FGM, Marques TA, De Barros M. Lipase properties, functions and food applications. Microb Enzym Technol Food Appl. 2017;214-240.
Crossref - Jaeger K-E, Dijkstra BW, Reetz MT. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnological Applications of Lipases. Annu Rev Microbiol. 1999;53(1):315-351.
Crossref - Wang Y, Srivastava KC, Shen GJ, Wang HY. Thermostable alkaline lipase from a newly isolated thermophilic Bacillus, strain A30-1 (ATCC 53841). J Ferment Bioeng. 1995;79(5):433-438.
Crossref - Sztajer H, Maliszewska I, Wieczorek J. Production of exogenous lipases by bacteria, fungi, and actinomycetes. Enzyme Microb Technol. 1988;10(8):492-497.
Crossref - Gupta PK. Soil, Plant, Water and Fertilizer Analysis. 2004:366. https://books.google.lk/books?id=xAKsnAEACAAJ
- Thakur S. Lipases, its sources, properties and applications: A review. Int J Sci Eng Res. 2012;3(7):1-29.
- Ghosh PK, Saxena RK, Gupta R, Yadav RP, Davidson S. Microbial lipases: production and applications. Sci Prog. 1996;79 ( Pt 2)(2):119-157.
- Heravi KM, Eftekhar F, Yakhchali B, Tabandeh F. Isolation and identification of a lipase producing Bacillus sp. from soil. Pakistan J Biol Sci. 2008;11(5):740-745.
Crossref - Patel P, Desai B, Science A, License A. Research Article Isolation, Identification and Production of Lipase Producing Bacteria from Oil Contaminated Soil. 2018;4(1):1-7.
- Reza KM, Ashrafalsadat N, Reza RM. Isolation and molecular identification of extracellular lipase-producing bacillus species from soil. 2014;5(1):132-139.
- Singh D, Kumar R, Sharma A, Kumar A. Lipase from Bacillus pumilus RK31: Production, Purification and Some Properties. World Appl Sci J. 2012;16(7):940-948. https://pdfs.semanticscholar.org/8583/33920f09208c4ae4d8c79febf194a04ea17c.pdf.
- Elibol M, Ozer D. Influence of oxygen transfer on lipase production by Rhizopus arrhizus. Process Biochem. 2000;36(4):325-329.
Crossref - Beisson F, Tiss A, Rivière C, Verger R. Methods for lipase detection and assay: a critical review. Eur J Lipid Sci Technol. 2000;102(2):133-153.
Crossref - Shimada Y, Sugihara A, Nagao T, Tominaga Y. Induction of Geotrichum candidum lipase by long-chain fatty acids. J Ferment Bioeng. 1992;74(2):77-80.
Crossref - Wang D, Wang J, Wang B, Yu H. A new and efficient colorimetric high-throughput screening method for triacylglycerol lipase directed evolution. J Mol Catal B Enzym. 2012;82:18-23.
Crossref - Sangeetha R, Geetha A, Arulpandi I. Optimization of protease and lipase production by Bacillus pumilus SG 2 isolated from an industrial effluent. Internet J Microbiol. 2012;5(2):1-8.
Crossref - Sagar K, Bashir Y, Phukan MM, Konwar BK. Isolation of Lipolytic Bacteria from Waste Contaminated Soil: A Study with Regard to Process Optimization for Lipase. Int J Sci Technol Res. 2013;2(10):214-218.
- Hasan F, Shah AA, Hameed A. Methods for detection and characterization of lipases: A comprehensive review. Biotechnol Adv. 2009;27(6):782-798.
Crossref - Ertuǧrul S, Dönmez G, Takaç S. Isolation of lipase producing Bacillus sp. from olive mill wastewater and improving its enzyme activity. J Hazard Mater. 2007;149(3):720-724.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.