Envelope protein of human coronavirus play significant role in an evolution and mutation of the virus life cycle. In the present research, the author evaluated amino acid sequences, its abundance and GC content of the genes that corresponds to envelope proteins’ of the human coronaviruses, identified from year 2003-2019. It includes SARS-CoV (2003), HCoV-NL-63 (2003), HCoV-HKU-1 (2004), MERS-CoV (2013), and SARS-CoV-2 (2019). The present research findings illustrated a point mutation in D2 location of SARS-CoV-2 (2019), representing Arginine in the place of Glutamic acid (SARS-CoV, 2003) where glycine was found deleted. SARS-CoV-2 (2019) coronavirus revealed increased abundance of Glutamic acid (100%), Asparagine (67%), Serine (300%) and Valine (85.7%) in comparison with HCoV-NL-63 (2003). We observed lower GC content in the SARS-CoV-2 (2019) among SARS-CoV (2003) and MERS-CoV (2013). The present findings have evolutionary significance and indicate SARS-CoV-2 (2019) adaptation in human.
Envelope Protein, Coronaviruses, SARS-CoV-2 (2019), Genetic evolution, GC content, Amino acids, Adaptation
The pandemic of SARS CoV-2 has spread across the globe in less than six months1. Family Coronaviridae is comprised of many pedigrees, identified in humans and birds worldwide2. In the human coronavirus category, SARS-CoV (2003), MERS-CoV (2013), and SARS-CoV-2 (2019) are extensively reported and studied strains3. Current SARS-CoV-2 (2019) strain possess human to human pathogenicity4. Due to adaptation in human host, a non-symptomatic recurrent mutation in SARS-CoV-2 (2019) study demonstrates 80% changes in the protein variety and at adaptation level. The envelope proteins are essential and significant functionality in viral life cycle including: protection of the genetic material, evading from host immune system and virus entry into the host cell5.
Genetic mutations are one of the most regular mechanism in the evolutionary cycle of viruses6. The present research is aimed to evaluate the genetic mutation in envelope protein genes and its evolutionary significance in human coronaviruses; identified from year 2003-2019. It includes SARS-CoV (2003), HCoV-NL-63 (2003), HCoV-HKU-1 (2004), MERS-CoV (2013), and SARS-CoV-27. Author studied the genetic mutation in envelope protein genes and the corresponding amino acid sequences of the selected viral strains. The genome sequence of selected coronaviruses was collected from NCBI [https:// www.ncbi.nlm.nih.gov/ nucleotide], respective amino acid sequences were aligned and evaluated in clustalW2 software8.
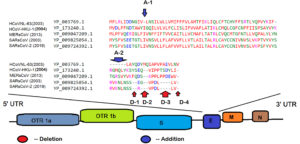
Phan1 reported presence of open reading frames (ORF) 1a and ORF1b in envelope protein genes of human coronaviruses followed by detection of spike, envelope, matrix and nucleocapside polyproteins. The present comparison of envelope proteins’ amino acid sequences was performed with oldest reported human coronavirus, HCoV-NL63 (2003). The alignments of all five coronaviruses envelope proteins’ amino acid sequences exemplified the addition and deletion of amino acids as represented in Fig. 1. A detail of amino acids addition and or deletion is represented in Table 1.
Table (1):
Addition and Deletion Pattern of Amino Acids in Envelope Proteins’ of Human Coronaviruses.
| Name of Human Coronaviruses | Additions of Amino Acids | Deletion of Amino Acids | ||||
|---|---|---|---|---|---|---|
| Location 1 (A-1) | Location 2 (A-2) | Location 1 (D-1) | Location 2 (D-2) | Location 3 (D-3) | Location 4 (D-4) | |
| HCoVNL-63(2003) | – | – | YQ | YMQ | PAEV | V |
| HCoV-HKU-1 (2004) | I | KRGMQLY | YS | Q – – | TSDY | – |
| MERsCoV (2013) | I | NTGRSVY | FG | S – – | PPDE | – |
| SARsCoV (2003) | I | SRVKNLN | – – | G – – | – – – – | – |
| SARsCoV-2 (2019) | I | SRVKNLN | – – | – – – | – – – – | – |
Fig. 1. Multiple Sequence Analysis of Amino Acid Sequence of Envelope Proteins’ of Human Coronaviruses
Briefly, a comparative analysis of SARS-CoV (2003) and SARS-CoV-2 (2019) illustrated deletion of glycine at D2 location in SARS-CoV-2 (2019) (Fig. 1) wherein the point mutation favored the replacement of glutamic acid (E) with Arginine (R). Further, we evaluated percent identity of the amino acid sequences of selected human coronaviruses, using percent identity matrix (data not shown). The author used clustalW2 software to infer the same8.
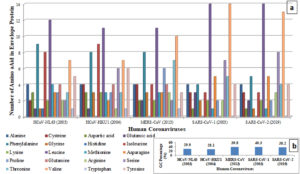
The present analysis revealed 96% sequence similarity of the envelope proteins’ amino acids among SARS-CoV (2003) and SARSCoV-2 (2019) coronaviruses. In contrast, SARSCoV-2 (2019) depicted 18.57%, 23.33% and 36.00% similarity index with HCoV-NL-63 (2003), HCoV-HKU-1 (2004) and MERS-CoV (2013) coronaviruses respectively. In addition, we studied the amino acid abundance of selected human coronaviruses using Expasy-protparam software. Comparative analysis among HCoV-NL-63 (2003) and SARS-CoV-2 (2019) coronaviruses revealed increased abundance of Glutamic acid (100%), Asparagine (67%), Serine (300%) and Valine (85.7%) in SARS-CoV-2 (2019) wherein a decrease of Aspartic acid (-66.6%), Histidine (-100%), Glutamine (-100%) and tryptophan (-100%) was observed (Fig. 2a).
Fig. 2. Abundance of Amino Acids Envelope Protein (a)and GC percentage of Envelope Protein gene sequence(b) among Human Coronaviruses
The author also performed the GC content analysis of envelope proteins’ genes among selected coronaviruses using Genomics % G-C content Calculator software. The analysis revealed that; SARS-CoV-2 (2019) had lower GC content in comparison with SARS-CoV (2003) and MERS-CoV (2013) (Fig. 2b).
Li et al9 reported correlation between low GC content of human coronaviruses and their adaptation in human lungs. Similarly, Schoeman et al10 reported correlation between envelope protein evolution among human coronaviruses and their pathogenicity, budding and envelope formation in the host. After year 2003; human coronaviruses have not been evaluated based on evolution of amino acid sequence, their abundance and GC content that corresponds to the envelope proteins. Taking into consideration the current pandemic of SARS-CoV-2 (2019) coronavirus, the progressive structural changes in amino acid sequence, their abundance and the GC content of envelope proteins’ should be determined urgently in order to investigate their contribution in assigning the host pathogenicity and interactions. Majorly, their evolutionary significance should be discovered, that would justify their adaptation functionalities and the target we can be sure of towards healing the SARS-CoV-2 (2019) coronavirus pandemic in the coming future.
ACKNOWLEDGMENTS
I would like to express my heartfelt thanks to Dr. Durga Aphale (Praj Matrix R&D Center, Division of Praj Industries Ltd, Pune.) for providing extended help on English editing of the manuscript
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
Viral gene sequences are available in ‘National Center for Biological Information’ (NCBI) server under the NCBI reference sequence numberYP_003769.1: HCoVNL-63(2003), YP_173240.1: HCoV-HKU-1 (2004), YP_009047209.1: MERsCoV (2013), YP_009825054.1: SARsCoV (2003) and YP_009724392.1: SARsCoV-2 (2019) respectively.
- Phan T. Novel coronavirus: From discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211.
Crossref - Shaw LP, Wang AD, Dylus D, et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol Ecol. 2020;29(17):3361-3379.
Crossref - De Souza Silva GA, Da Silva SP, Da Costa MAS, et al. SARS-CoV, MERS-CoV and SARS-CoV-2 infections in pregnancy and fetal development. J Gynecol Obstet Hum Reprod. 2020;49(10):101846.
Crossref - Nishiura H, Linton NM, Akhmetzhanov AR. Initial Cluster of Novel Coronavirus (2019-nCoV) Infections in Wuhan, China Is Consistent with Substantial Human-to-Human Transmission. J Clin Med. 2020;9(2):488.
Crossref - Buchmann JP, Holmes EC. Cell Walls and the Convergent Evolution of the Viral Envelope. Microbiol Mol Biol Rev. 2015;79(4):403-418.
Crossref - Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6(7):e1001005.
Crossref - Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-733.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Li Y, Yang X, Wang N, et al. GC usage of SARS-CoV-2 genes might adapt to the environment of human lung expressed genes. Mol Genet Genomics. 2020;295(6):1537-1546.
Crossref - Schoeman D, Fielding BC. Coronavirus envelope protein: Current knowledge. Virol J. 2019;16(1):69.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.