ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus is one of the major causes of life threatening pneumonia, especially in immunocompromised population. In HIV positive patients, S. aureus associated pneumonia can be either health care associated or community acquired and responsible for high rate of mortality. In this study total 102 throat swab samples of HIV-Infected Patients with suspected pneumonia were collected during 2014-2016, out of them 46 samples (45.1%) were found positive for S. aureus by biochemical tests. 38 (82.6%) isolates were found multiple drug resistant while 9 (19.6%) strains showed resistance to cefoxitin antibiotic, were considered as methicillin resistant Staphylococcus aureus (MRSA). Only one strain (2%) was found vancomycin intermediate (VISA), remaining 98% isolates were sensitive to vancomycin antibiotic. In PCR test, all cefoxitin resistant strains were found positive for the presence of MecA gene. Biofilm former S. aureus were screened by tissue culture plat (TCP) methods. In TCP assay, 21 (45.6 %) isolates were confirmed as high biofilm formers (OD value > 0.250), 16 (34.8 %) were moderate biofilm formers (OD values- between 0.150 to 0.250), while 9 (19.6 %) were low biofilm formers (OD value < 0.150). A significant association was found among multiple drug resistance and high biofilm formation (p value < 0.05). High prevalence of biofilm forming MDR isolates in airways of pneumonia suspected HIV patients is matter of great concern as poor antibiotic response may cause more severe diseases with increasing cost and duration of treatment. The MecA gene might be a cause of methicillin resistance among MRSA isolates.
Multiple Drug Resistance, Biofilm, MRSA, Pneumonia, Immunocompromise, HIV
Opportunistic pneumonias are the major cause of HIV-associated severe respiratory illness with high rate of mortality1. The opportunistic pneumonias among HIV patients may include mycobacterial, viral, bacterial, parasitic and fungal pneumonias, among them bacterial pneumonia is the most common type of opportunistic pneumonia2-4. In HIV positive persons, the rate of bacterial pneumonia is higher than persons without HIV3,5. The clinical characteristics of pneumonia are fever, chills, chest pain, shortness of breath etc. Although bacterial pneumonia can occur at any stage of HIV infection but the incidence of bacterial pneumonia increases with declines in CD4 cell count1. Most common bacterial pathogens associated with community acquired pneumonia during HIV infection are Streptococcus pneumoniae, Haemophilus species and Staphylococcus aureus6.
In contrast, S. aureus normally colonize in skin and mucous membranes in the upper respiratory tract of healthy individuals without any complications, but in immunocompropised people, S. aureus has been associated with many syndromes such as pneumonia, bacteraemia, septicaemia, urinary tract infections, diarrhea and skin infections with increased severity7. The detection and characterization of S. aureus among clinical samples is generally done by biochemical tests such as mennitol fermentation, coagulase and DNase. Few studies suggested the polymerized chain reaction (PCR) as molecular technique using amplification of nuc gene for early diagnosis of S. aureus8.
The prevalence of multiple antibiotic resistances in S. aureus is increasing globally, which causes poor antibiotic response and economical burden especially in developing countries7. In the 1960s, first case associated with methicillin resistant S. aureus (MRSA) was reported and in the late 1990s It became a well known cause of community and hospital associated infections9. Previous studies suggested that resistance to methicillin among S. aureus is associated with presence of mecA gene which encodes penicillin binding protein 2a (PBP 2a). Staphylococcal chromosome cassette mec (SCCmec), a mobile genetic element of 21 to 60 kb contains mecA gene. SCCmec may also carry genes such as pT181, Tn554 and pUB110 which may associated with resistance for non-beta lactam class of antibiotics10. Expression of PBPs provides capacity to bacterial cells to continue cell wall synthesis in presence of higher concentration of cell wall synthesis inhibitors11. Molecular detection of the highly conserved mecA gene using PCR amplification has been proved as a benchmark for the early diagnosis of MRSA in the clinical samples12. As an additional virulence factor many S. aureus strains have capacity to form biofilms on biotic and abiotic surfaces13. Biofilms are multilayered arrangement of bacterial cells enclosed in exopolysaccharide matrix, which helps bacterial cells in survival, providing protection against extreme environmental conditions, antimicrobial agents and host immunity14,15.Because of higher level of resistance against host immune responses and antibiotics, biofilm forming S. aureus may lead to sever and chronic disease in individuals with weak immunity such as HIV positive patients. Although in HAART era, a significant fall in opportunistic infections has been noticed, but HIV positive people are still at high risk of opportunistic bacterial respiratory infections16.
HIV patients are under high burden of medications, wrong selection of antibiotics for the treatment not only enhances duration of treatment and financial burden but also make disease more complicated and life threatening. Early detection of S. aureus and evaluation of their virulence such as antibiotic resistance, presence of mecA gene and biofilm formation ability is essential for selection of proper antibiotic for the treatment of pneumonia in HIV patients. The involvement of biofilm forming, MRSA in HIV patients suffering from pneumonia, is not well studied. The aim of this study is to detect the prevalence of multiple drug resistant and biofilm forming Staphylococcus aureus among pneumonia suspected HIV patients and evaluate the presence of mecA gene among isolates.
Isolation and characterization of S. aureus from clinical samples
The isolation and characterization of S.aureus from clinical samples (throat swab) was done by direct throat swab inoculation on mannitol salt agar (MSA) (HiMedia laboratories private limited, India) plates, followed by incubation at 37 °C for 24 h. Gram staining was performed followed by biochemical confirmation tests such as DNase, mannitol fermentation and coagulase tests. All strains which were positive for mannitol fermentation, coagulase and DNase tests were confirmed as Staphylococcus aureus17.
Antibiotic sensitivity and detection of MRSA by disc diffusion method
The antibiotic sensitivity test was performed by modified Kirby-Bauer disc diffusion method using Mueller-Hinton agar (HiMedia laboratories private limited, India) for the antibiotic discs- azithromycin (25 µg), chloramphenicol (30 µg), cefoxitin (30 µg), clindamycin (2 µg), sulfamethoxazole/trimethoprim (25 µg), gentamycin (10 µg), levofloxacin ( 5 µg), linzolid (30 µg), vancomycin (30 µg) following Clinical and Laboratory Standards Institute (CLSI, 2013) guidelines 18. All cefoxitin resistant strains were confirmed as MRSA.
Bacterial DNA Isolation
Bacterial log culture was prepared in tripticase soy broth (HiMedia laboratories private limited, India). 250 µL of broth culture was centrifuged at 5000 rpm for 10 min to settle bacterial cells. Pellet was resuspended in 50 µL of dd water, followed by addition of 5 µL lysostaphin solution (Sigma-Aldrich Corp.) and incubated at 37 °C for 30 min, followed by DNA extraction processes using PureLinkTM genomic DNA mini Kit (Thermo Fisher Scientific) according to manufacturer’s instructions. Extracted bacterial DNA was stored at – 20 °C.
Detection of nuc and mecA genes by PCR amplification
A multiplex PCR was standardized for screening of nuc and mecA genes among isolates, using specific primers (Table-1). The thermal cycling conditions: an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 Sec, annealing at 51 °C for 30 Sec and extension at 72 °C for 90 Sec, final extension at 72 °C for 10 min., followed by agarose gel electrophoresis. DNA bands were visualized under gel imaging system (Bio-Rad Laboratories).
Table (1):
Primers used for the molecular detection of nuc and mecA genes.
Primers |
Sequences of primers |
Size of amplified products (bp) |
|---|---|---|
Nuc-F Nuc-R |
5 -GCGATTGATGGTGATACGGTT-3 5 -AGCCAAGCCTTGACGAACTAAAGC-3 |
367 bp |
Mec-F Mec-R |
5’-GTT GTA GTT GTC GGG TTT GG-3’ 5’CTT CCA CAT ACC ATC TTC TTT AAC-3’ |
533 bp |
Detection of biofilm forming S. aureus
Screening of biofilm forming S. aureus strains was done by tissue culture plate (TCP) method19 with some modification20. TSB media (HiMedia laboratories private limited, India) enriched with 1% glucose was inoculated with isolates and incubated for 24 h at 37 °C. Log cultures were diluted (1:100) with fresh TSB medium, each well of 96 welled TCP was filled with inoculated media (200 µL) followed by incubation for 24 h at 37 °C. After incubation the content of plate was removed and washed three times with 200 µL of phosphate buffer saline (PBS). 0.25% crystal violet solution (200 µL) was added to each well and incubated for 20 min and rinse thrice with PBS. 200 µL of 95% ethyl alcohol was added. Optical density (OD) of stained biofilm in each well was determined by ELISA Reader (Thermo Fisher Scientific, India) at wavelength 630 nm. TCP assay was repeated three times for each strain.
Statistical analysis
Statistical analysis was performed using SPSS software (IBM SPSS statistics 20). The chi square test was performed to compare MDR and non MDR isolates for the presence of virulence factors such as biofilm formation and mecA gene.
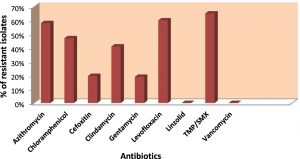
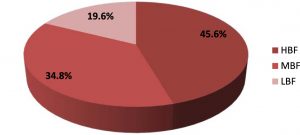
In this study total 102 throat swab samples of pneumonea suspected HIV patients were collected during 2014-2016. 46 samples (45.1%) were found positive for S. aureus by biochemical tests. In antibiotic sensitivity test, highest 65% of isolates were found resistant for antibiotic sulfamethoxazole/trimethoprim followed by levofloxacin 60%, azithromycin 58%, chloramphenicol 47%, clindamycin 41%, cefoxitin 20%, gentamycin 19%. Resistance for vancomycin and linzolid antibiotics was not found among isolates (fig. 1), while only one isolate (2%) was found vancomycin intermediate S. aureus (VISA).Out of total confirmed S. aureus, 38 (82.6%) isolates showed resistance for three or more antibiotics in antibiotic sensitivity test, considered as MDR, while 9 (19.6%) isolates showed resistance for cefoxitine antibiotic were confirmed as MRSA. All MRSA were found MDR. In PCR test, all cofirmed S. aureus showed amplification of nuc gene while all MRSA strains were found positive for presence of mecA gene (fig. 2). Biofilm forming S. aureus strains were screened by tissue culture plat (TCP) method (fig. 3). In TCP assay, 21 (45.6 %) isolates were confirmed as high biofilm forming isolates (OD value > 0.250), 16 (34.8 %) were moderate biofilm forming (OD values- between 0.150 to 0.250) while 9 (19.6 %) were low biofilm formers (OD value < 0.150) (fig. 4). A significant association was found among multiple drug resistance and high biofilm formation (p value< 0.05) (Table 2).
Table (2):
Association of bacterial virulence factors (biofilm formation and presence of mecA gene) with antibiotic sensitivity.
| S.N. | Bacterial virulence Factors |
Staphylococcus aureus Isolates (N=46) | p values* | |
|---|---|---|---|---|
| MDR isolates (N=38) |
Non MDR isolates (N=8) |
|||
| 1. | Biofilm formation HBF MBF LBF |
20 (95%) 13 (81%) 5 (55%) |
1 (5%) 3 (19%) 4 (45%) |
0.03* 0.9 0.017* |
| 2. | Presence of MecA Gene | 9 (100%) | 0 (0%) | 0.125 |
*Chi-square test was performed to compare the bacterial virulence factors (Biofilm formation, Methicillin resistance, Presence of MecA gene) between MDR and non MDR S. aureus (p-value <0.05 was considered significant).
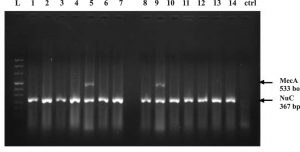
Fig. 2. Agarose gel images; Showing PCR amplification of specific Nuc (367 bp) and MecA (533 bp) genes of Staphylococcus aureus isolates: Lane- L is DNA Ledder (1000bp), Lane 1 to 14 – PCR amplification of Nuc gene, Lane 5, 9 – PCR amplification of MecA gene. Lane- ctrl is negative control.
Fig. 4. Percentage of biofilm forming Staphylococcus aureus isolates; OD values less than 0.150 were recorded as low biofilm forming isolates, those with OD values between 0.150 to 0.250 were considered as moderate biofilm formers and those with OD values above 0.250, were considered as high biofilm forming isolates.
Acquired immunodeficiency syndrome (AIDS) is a major health issue specially in developing world7. Weak immunity in HIV positive people place them at increased risk of opportunistic infections. Opportunistic associated respiratory illness is more common in immunocompromised people than healthy individuals3,5. Although most of the respiratory infection are self limiting and can be treated with medications but in HIV patients these infection may cause severe complications and life threatening conditions1. In HAART era the immunity status of HIV patients has significantly improved due to improvement in the level of CD4 cell count but the threat of opportunistic infections among HIV positive individuals cannot be denied16.
S. aureus is a common opportunistic pathogen associated with community acquired as well as nosocomial infections7. The emergence of antibiotic resistance and rise in level of virulence among S. aureus is a burning health issue both within the community and hospital settings. Occurrence of MRSA in respiratory infections has increased over time among HIV patients21, 22. MRSA associated pneumonia among HIV patients can be community acquired or health-care associated and may lead to higher rate of morbidity and mortality. The rate of colonization by S. aureus is higher in HIV positive patients. As a colonizer MRSA may associated with increased risk of infections23, 24. In our study, out of 102 throat swab samples of HIV patients, 46 samples (45%) were positive for S. aureus. Out of the confirmed S. aureus, 82.6% were MDR while 19.6% were found MRSA. All MRSA were MDR. In a previous study, S. aureus was found associated with 25% of pneumonia cases in HIV-positive patients, among them 65% were MRSA25. Another study showed 85.7% prevalence of MRSA in cutaneous abscesses of HIV patients26.
In antibiotic sensitivity test, the highest resistance was noticed for sulfamethoxazole/ trimethoprim antibiotic, while no resistance was found for vancomycin and linzolid antibiotic. A previous study supports our results, showing no resistance in MRSA for teicoplanin, vancomycin, and linezolid antibiotics27. In some other reports, MRSA were completely found sensitive for vancomycin 28 and linzolid antibiotics 29, 30. While in another study high resistance for vancomycin was observed and sulfamethoxazole/trimethoprim (TMP/SMX) antibiotic found effective against pathogenic S. aureus strains26, showing dissimilarity to our data.
The biofilm formation among S. aureus is a known virulence factor13. It is estimated that bacterial biofilms are associated with 65% of all bacterial infections. The involvement of biofilms found in both, device and non-device associated infections31. In our study 45.6 % isolates were found high biofilm formers, 34.8 % moderate biofilm former while 19.6 % were low biofilm formers. In a study 48 % biofilm forming S. aureus isolates were reported in different clinical samples of HIV positive patients. In our study, a significant association among high biofilm formation with multiple antibiotic resistance was observed. Similarly, Samie et al., correlated biofilm formation with b-lactamase production, out of the 14 strong biofilm formers, 9 (64.3%) were found b-lactamase positive7. However, the association of biofilm forming and multiple drug resistant S. aureus in pneumonia among HIV positive patients is not well studied. Higher level of drug resistance and biofilm formation among isolates, may lead to sever pneumonia, especially in immunocompromised patients. Early detection of S. aureus and evaluation of their virulence such as antibiotic resistance, presence of mecA gene and biofilm formation is essential for selection of proper antibiotic for the treatment of pneumonia in HIV patients.
High prevalence of biofilm forming MDR isolates in airways of HIV-Infected Patients with suspected pneumonia is matter of great concern as poor antibiotic response may cause more severe diseases with increasing cost and duration of treatment. The methicillin resistance among MRSA isolates showed positive association with the presence of mecA gene. Biofilm formation among S. aureus isolates might be a cause of increasing multiple antibiotic resistance.
Acknowledgements
We would like to thank to Indian council of medical research (ICMR) for financial supports.
- Huang, L., Crothers, K.A.. HIV-associated opportunistic pneumonias. Respirology., 2009; 14(4): 474–485.
- Jones, J.L., Hanson, D.L., Dworkin, M.S., Alderton, D.L., Fleming, P.L., Kaplan, J.E., et al. Surveillance for AIDS-defining opportunistic illnesses, 1992-1997. MMWR CDC Surveill Summ., 1999; 48(2): 1–22.
- Hirschtick, R.E., Glassroth, J., Jordan, M.C., Wilcosky, T.C., Wallace, J.M., Kvale, P.A., et al. Bacterial pneumonia in persons infected with the human immunodeficiency virus. Pulmonary Complications of HIV Infection Study Group. N Engl J Med., 1995; 333(13): 845–51.
- Salomon, N., Gomez, T., Perlman, D.C., Laya, L., Eber, C., Mildvan, D. Clinical features and outcomes of HIV-related cytomegalovirus pneumonia. Aids., 1997; 11(3): 319–24.
- Falco, V., Sevilla, F.D., Alegre, J., Barbe, J., Ferrer, A., Ocana, I., Ribera, E. and Vazquez, J.M. Bacterial pneumonia in HIV-infected patients: a prospective study of 68 episodes. European Respiratory Journal., 1994; 7: 235-239.
- Burack, J.H., Hahn, J.A., Saint-Maurice, D., Jacobson, M.A. Microbiology of community-acquired bacterial pneumonia in persons with and at risk for human immunodeficiency virus type 1 infection. Implications for rational empiric antibiotic therapy. Arch Intern Med., 1994; 154(22): 2589–96.
- Samie, A. and Shivambu, N.A. Biofilm production and antibiotic susceptibility profiles of Staphylococcus aureus isolated from HIV and AIDS patients in the Limpopo Province, South Africa. African Journal of Biotechnology., 2011; 10(65): 14625-14636.
- Brakstad, O.G., Aasbakk, K. and Maeland, J.A. Detection of Staphylococcus aureus by Polymerase Chain Reaction Amplification of the nuc Gene Journal Of Clinical Microbiology., 1992; 30: 1654-1660.
- Hidron, A.I., Kempker, R., Moanna, A., Rimland, D. Methicillin-resistant Staphylococcus aureus in HIV-infected patients. Infection and Drug Resistance., 2010; 3: 73–86.
- Wielders, C.L.C., Fluit, A.C., Brisse, S., Verhoef, J. and Schmitz F.J. MecA Gene is widely disseminated in Staphylococcus aureus population. Journal of clinical microbiology., 2002; 40: 3970–3975.
- Aedo, S., Tomasz, A. Role of the Stringent Stress Response in the Antibiotic Resistance Phenotype of Methicillin-Resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy., 2016; 60: 2311- 2317.
- Al-Abbas, M.A. Antimicrobial susceptibility of Enterococcus faecalis and a novel Plano-microbium isolate of bacteremia. International Journal of Medicine and Medical Sciences., 2012; 4: 19–27.
- Eftekhar, F. and Dadaei, T. Biofilm Formation and Detection of IcaAB Genes in Clinical Isolates of Methicillin Resistant Staphylococcus aureus. Iranian Journal of Basic Medical Sciences., 2011; 14: 132-136.
- O’Gara, J.P. and Humphreys, H. Staphylococcus epidermidis biofilms importance and implications. H Med Microbiol., 2001; 50: 582-87.
- Gad, G.F.M., El-Feky, M.A., El-Rehewy, M.S. and Hassan, M.A. et al. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. Infect Dev Ctries., 2009; 3: 342-351.
- Lopez, P.C., Martin, Z.M., Benitez, E., Fernandez, G.C., Guerrero, F., Rodriguez, I.M. and Giron G.J.A. Pneumonia in HIV-infected patients in the HAART era: incidence, risk, and impact of the pneumococcal vaccination. J Med Virol., 2004; 72: 517-24.
- Rahimi, F., Bouzari, M., Katouli, M. and Pourshafie M.R. Antibiotic resistance pattern of methicillin-resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbiol., 2013; 6: 144-149.
- Clinical and Laboratory Standards Institute, “Performance standards for antimicrobial susceptibility testing: twenty third informational supplement edition,” CLSI Document M100-S23, CLSI, Wayne, Pa, USA, 2013.
- Christensen, G.D., Simpson, W.A., Younger, J.A., Baddour, L.M., Barrett, F.F., Melton, D.M. and Beachey, E.H. Adherence of coagulase negative Staphylococci to plastic tissue cultures: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol., 1985; 22: 996-1006.
- O’Toole, A.G. and Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescence WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular microbiology., 1998; 28: 449.
- Skiest, D., Brown, K. and Hester, J., et al. Community-onset methicillin resistant Staphylococcus aureus in an urban HIV clinic. HIV Med., 2006; 7: 361–368.
- Crum, C.N., Weekes, J. and Bavaro, M. Recurrent community-associated methicillin-resistant Staphylococcus aureus infections among HIV infected persons: incidence and risk factors. AIDS Patient Care STDS., 2009; 23: 499–502.
- Kluytmans, J., Van, B.A., Verbrugh, H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev., 1997; 10: 505–20.
- Safdar, N., Bradley, E.A. The risk of infection after nasal colonization with Staphylococcus aureus. AmJ Med., 2008; 121(4): 310–5.
- Franzetti, F., Grassini, A., Piazza, M. Nosocomial bacterial pneumonia in HIV-infected patients: risk factors for adverse outcome and implications for rational empiric antibiotic therapy. Infection., 2006; 34(1): 9–16.
- Krucke, G.W., Grimes, D.E., Grimes, R.M. and Dang, T.D. Antibiotic resistance in the Staphylococcus aureus containing cutaneous abscesses of HIV patients. Am J Emerg Med., 2009; 27(3): 344–347.
- Khan, R.A., Rahman, A.U., Ahmad A., Jaseem, M., Jabbar, A., et al. Prevalence and Antibiotic Susceptibility Profile of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Different Clinical Samples in District Peshawar. J. Appl. Environ. Biol. Sci., 2014; 4: 40-46.
- Maple, P.A.C., Hamilton-Miller, J.M.T. and Brumfitt, W., World-wide antibiotic resistance in methicillin-resistant Staphylococcus aureus. The Lancet., 1989; 333: 537-540.
- Stratchounski, L.S., et al. Antimicrobial resistance of nosocomial strains of Staphylococcus aureus in Russia: results of a prospective study. Journal of chemotherapy., 2005; 17: 54-60.
- Anbumani, N., Kalyani, J., Mallika, M. Prevalence of Methicillin-Resistant Staphylococcus aureus in a Tertiary Referral Hospital in Chennai, South India. Blood., 2006; 126: 38.
- Lewis K. Riddle of Biofilm Resistance. Antimicrob Agents Chemother., 2001; 45(4): 999–1007.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.