ISSN: 0973-7510
E-ISSN: 2581-690X
Neonatal sepsis due to carbapenem-resistant Klebsiella pneumoniae (CRKP) is a global cause of morbidity and mortality, especially in East and Southeast Asia. Among different sources of CRKP implicated in sepsis, neonatal skin and gut colonization are important reservoirs. Despite being endemic for CRKP infections, there is an extreme paucity of data on CRKP colonization in neonates. Therefore, this prospective cohort study was performed to detect the frequency of gut colonization with CRKP in the neonates following admission in the neonatal unit of a tertiary care hospital in India. The study was conducted in the 14 bedded neonatal unit and infection control section. Surface swabs from skin and rectal swabs were cultured for bacterial identification from every neonate at admission and at 24 h, 48 h, and 72 h. Isolates were genotypically tested for phylogeny groups, screened for carbapenem resistance by imipenem E-test and typed by Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR. Data for the CRKP colonized and non-colonized neonates were statistically analyzed. A total of 30 neonates and 180 swabs were studied. Gram-positive cocci in skin, whereas for rectal colonization, K. pneumoniae (genotype K. pneumoniae subsp. pneumoniae) was the commonest. The frequency of CRKP colonization was 33%, with 20% colonization pressure at admission. All the CRKP had blaNDM gene. On ERIC-PCR, multiple clones were seen. The study highlighted the high burden of blaNDM carrying CRKP colonization in neonates at admission, thus emphasizing the urgent need for the implementation of stringent infection control policies to prevent nosocomial transmission of these colonizers.
Rectal, Skin, Admission, blaNDM, NICU
In developing countries including those of Africa, South-east Asia and East Asia, Klebsiella pneumoniae has been the predominant pathogen for sepsis among neonates.1 Any neonatal unit typically consists of its own specific signatory microflora, which is different from other common-built hospital environments.2,3 Often, this microbiome of the neonatal intensive care unit (NICU) environment is reflected in the gut colonization of the neonates. Colonized organisms are usually specific with ability to persist by tolerating several environmental stresses like selective pressures of disinfectants used in surface cleaning2 and antibiotics3 used in the NICU. In this relevance, the neonatal unit of the tertiary care hospital in the present study has previously experienced several well controlled small outbreaks of K. pneumoniae associated neonatal sepsis along with few major outbreaks with extensively drug-resistant K. pneumoniae.4-6 In most of these outbreaks and in others, environmental reservoir has been studied to be the most common source of contamination.7 Carbapenem-resistant K. pneumoniae (CRKP), itself being one among the “critical pathogen of utmost priority’’ labelled by World Health Organization (WHO) and the Indian Priority Pathogen List (IPPL), is being recently encountered with increased frequency in neonatal sepsis all over the globe.8,9 Several sources of CRKP have been implicated in neonatal sepsis. Among these, contamination of hands of healthcare workers, neonatal skin and gut colonization and maternal gut colonization are the commonly studied sources.6,10-12 In context of gut colonization, there have been previous studies supporting as well as not supporting the association between gut/rectal colonization with CRKP in pediatric population with infections due to CRKP.13,14 Being in an endemic set-up for CRKP infections, this study was performed to detect the frequency of gut colonization with CRKP in the neonates following admission in the NICU and their subsequent course.
Study site and design
This was a single-centre, prospective, cohort study conducted over a period of 3 months in the 14 bedded NICU and infection control section of the Department of Microbiology of a tertiary care hospital in Varanasi. The study was approved by the Institute ethical committee (No. Dean/2021/EC/2598) and informed consent was obtained from the guardian of the neonates prior to sample collection.
Study samples
Surface swabs (HiMedia Laboratories Pvt. Ltd., India) from skin of the axilla, around the umbilicus and abdominal surface along with rectal swabs were collected from every neonate at admission in the NICU. Repeat swabs were collected from the same neonates at 48 h and 72 h. Those neonates were included in the study with all 3 sets of swabs appropriately collected and also having complete demographic data. Very sick neonates, those who did not survive more than 72 h post-admission, those with unidentified comorbidities and those where informed consent was not provided were excluded. Demographic details including age at admission, gender, body weight, mode of delivery and clinical details including presence and type of symptoms were noted from the NICU records. Maternal complications and final outcome of the neonate in the NICU were also noted. Data on empirical antibiotic coverage and the blood culture report was recorded. Admission positive was considered for those neonates with at least 1 screening sample positive within 24 h whereas acquired positive were those with positive screening samples at 48h or 72 h. Colonization pressure at admission was calculated by dividing the number of neonates colonized with CRKP with the total number of patients.
Processing of the samples in the laboratory
The swabs were immediately transported to the laboratory and inoculated onto blood agar and MacConkey agar (HiMedia Laboratories Pvt. Ltd., India). Following incubation organisms were identified based on conventional Gram’s staining and routine biochemical tests.15
Further screening for drug-resistant isolates was done as described. For screening of methicillin-resistant Staphylococcus aureus (MRSA), cefoxitin disc (30 µg) was used as per standard. CRKP were screened by disc diffusion against ertapenem (10 µg) and confirmed by imipenem E-test (HiMedia Laboratories Pvt Ltd, India). Vancomycin screen agar was used for Enterococci.16
Molecular characterization of CRKP isolates
All the isolates were tested for the presence of carbapenemase encoding genes. Briefly, multiplex PCR was performed for the amplification of class A (blaSME, blaNMC, blaGES, blaKPC), class B (blaIMP, blaVIM, blaNDM), and class D (blaOXA-48), carbapenemase genes using primers and reaction conditions as described elsewhere.17,18
Detection of clonality in CRKP by ERIC PCR
Classification of K. pneumoniae into three phylogeny groups K. pneumoniae subsp. pneumoniae (KpI), K. pneumoniae subsp. quasipneumoniae (KpII) and K. pneumoniae subsp. variicola (KpIII) was done by one-step multiplex PCR.19 The heterogeneity of the isolates was determined by enterobacterial repetitive intergenic consensus (ERIC) PCR using the universal primer ERIC-F (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-R (5′-AAGTAAGTGACTGGGGTGAGCG-3′). Amplification was done under previously described reaction conditions, and the bands patterns were analyzed by agarose gel electrophoresis.20 Based on the band patterns, the dendrogram was constructed by using computer program NTSYS-pc version 2.0.
Statistical analysis
Characteristics of the CRKP colonized and not colonized neonates were compared by Fisher’s exact test (Medcalc Statistical Software version 19.2.6, Ostend, Belgium). Colonization pressure at admission was calculated by dividing the number of neonates colonized with CRKP at admission by the total number of neonates.
A total of 30 neonates comprising of 19 males and 11 females were included in the study. The mean body weight of the neonates was 2259.81 ± 724.78 g. Mean age at admission was 3.73 ± 6.15 d. The different mode of delivery in these neonates were emergency lower segment Caesarean section (emLSCS) in 15 (50%), spontaneous vaginal delivery in 13 (43.33%) and elective LSCS in 2 (6.66%). Prematurity was seen in 8 (26.66%) of the neonates, while 11 (36.66%) presented with respiratory distress. The outcome in these neonates was 7 (23.33%) were discharged, 12 (40%) expired and 11 (36.66%) were shifted following response to treatment.
Table (1):
Distribution of organisms isolated from surface swabs from the skin of the neonates
| Organisms | Time of swab collection | Total | ||
|---|---|---|---|---|
| 24 h, n (%) | 48 h, n (%) | 72 h, n (%) | ||
| MSSA | 8 (25.00) | 5 (15.62) | 3 (9.37) | 16 (16.66) |
| MRSA | 5 (15.62) | 1 (3.12) | 6 (18.75) | 12 (12.50) |
| CoNS | 2 (6.25) | 3 (9.37) | 6 (18.75) | 11 (11.45) |
| Klebsiella pneumoniae | 2 (6.25) | 4 (12.50) | 5 (15.62) | 11 (11.45) |
| Acinetobacter spp. | 1 (3.12) | 2 (6.25) | 0 (0.00) | 3 (3.12) |
| Other Enterobacterales | 2 (6.25) | 0 (0.00) | 0 (0.00) | 2 (2.08) |
| Other Gram-positive cocci | 1 (3.12) | 2 (6.25) | 1 (3.12) | 4 (4.16) |
| Sterile | 11 (34.37) | 15 (46.87) | 11 (34.37) | 37 (38.54) |
MSSA= methicillin-susceptible, Staphylococcus aureus, MRSA= methicillin-resistant Staphylococcus aureus, CoNS= coagulase-negative staphylococci
Table (2):
Distribution of organisms isolated from rectal swabs of the neonates
| Organisms | Time of swab collection | Total | ||
|---|---|---|---|---|
| 24 h, n (%) | 48 h, n (%) | 72 h, n (%) | ||
| Klebsiella pneumoniae | 7 (23.33) | 13 (40.62) | 15 (46.87) | 35 (37.23) |
| MRSA | 1 (3.33) | 1 (3.12) | 0 (0.00) | 2 (2.12) |
| CoNS | 1 (3.33) | 0 (0.00) | 0 (0.00) | 1 (1.06) |
| MSSA | 0 (0.00) | 1 (3.12) | 1 (3.12) | 2 (2.12) |
| Enterococci | 1 (3.33) | 2 (6.25) | 1 (3.12) | 4 (4.25) |
| Other Enterobacterales | 0 (0.00) | 2 (6.25) | 0 (0.00) | 2 (2.12) |
| Pseudomonas spp. | 1 (3.33) | 0 (0.00) | 0 (0.00) | 1 (1.06) |
| Sterile | 19 (63.33) | 13 (40.62) | 15 (46.87) | 47 (49.99) |
MSSA= methicillin-susceptible Staphylococcus aureus, MRSA= methicillin-resistant Staphylococcus aureus, CoNS= coagulase-negative staphylococci
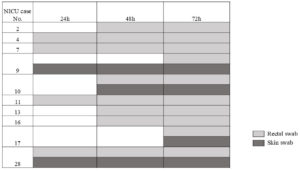
As per protocol, a total of 180 swabs were collected from which 106 organisms were isolated. The distribution of the organisms at 24 h, 48 h and 72 h collection has been shown in Table 1 for skin and Table 2 for rectal colonization. The most common colonizer in skin was Gram-positive cocci whereas for rectal colonization, K. pneumoniae was the commonest. Of the 30 neonates, 15 were colonized with K. pneumoniae of which 7 (46.66%) were admission positive and 8 (53.33%) were acquired positive. Of the 7 admission positive neonates, 4 (57.14%) had rectal colonization with CRKP. Of the acquired positive, 6 (75%) acquired rectal colonization with CRKP at 48 h and 2 (25%) at 72 h. In case of skin colonization, 2 (13.33%) were admission positive with K. pneumoniae both of which were CRKP. The colonization pressure of CRKP at admission was 20% (6 of 30). Of the acquired positive skin colonizers, 3 (75%) were positive for K. pneumoniae at 48 h of which 2 were CRKP and 1 at 72 h which was CRKP. Therefore, 33.33% neonatal CRKP colonization was detected (10) in the first 72 h of admission in the NICU. The distribution of the CRKP isolates among the 10 colonized neonates has been depicted in Figure 1. While rectal colonization predominated in the majority of the cases 5 (50%), simultaneous colonization of both skin and rectum was seen in 3 (30%) neonates and 1 (10%) neonates showed skin colonization with CRKP followed by rectal colonization. The characteristics of the CRKP colonized and not colonized neonates have been shown in Table 3. However, none of the neonates included in this study were infected with CRKP sepsis, though isolation of CRKP in blood of neonates (2 cases out of 54) were noted in this period. These neonates were not included in the study as they did not adhere to the inclusion criteria.
Table (3):
Comparison of characteristics of CRKP colonized positive and negative neonates
Characteristics |
Neonates with CRKP colonization, N = 10 |
Neonates without CRKP colonization, N = 20 |
p-value |
|---|---|---|---|
Mean age (days) |
1.40 ± 0.91 |
5.10 ± 7.35 |
0.13 |
Mean birth weight (grams) |
1993.75 ± 709.15 |
2386.11 ± 721.45 |
0.16 |
Mode of delivery: emLSCS |
6 (60.00) |
9 (45.00) |
0.61 |
SVD |
4 (40.00) |
9 (45.00) |
– |
Prematurity |
3 (30.00) |
5 (25.00) |
0.77 |
Low birth weight (
| 4 (40.00) |
8 (40.00) |
1.00 |
Associated comorbidities |
9 (90.00) |
15 (75.00) |
0.34 |
Maternal complications |
2 (20.00) |
12 (60.00) |
0.04* |
Sepsis |
6 (60.00) |
7 (35.00) |
0.19 |
Outcome (Expired) |
5 (50.00) |
7 (35.00) |
0.49 |
*p < 0.05, emLSCS= emergency lower segment Caesarean section, SVD= spontaneous vaginal delivery
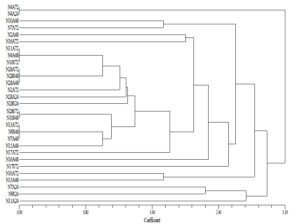
All the K. pneumoniae genotypes were identified as K. pneumoniae subsp. pneumoniae. All the CRKP harboured carbapenemase encoding blaNDM gene. On ERIC-PCR, multiple clones of CRKP were seen. However, similar clones were seen in skin and rectal colonization of each neonate, as shown in Figure 2.
The study highlights the high burden of CRKP colonization in neonates in a typical tertiary care NICU (33%), with 20% colonization pressure at admission. This study is important in the following aspects. There has been extreme paucity of data on CRKP colonization especially in neonatal population from East and Southeast Asia, which are endemic for CRKP infections. A study from Vietnam revealed 21% admission positive CRKP and 74% acquired CRKP, with subsequent CRKP infection in carrier group (45.9%) being significantly higher than that of the baseline non-KP carrier group 8.6%.21 A study from Turkey showed 2.6% rectal colonization with CRKP in NICU and infection rate of 18% among the carriers.13 On the other hand, another study on pediatric population had reported lower rate of infections of 3.4% among 29.5% colonized patients.14 This study adds to the existing data on the burden of CRKP colonization, thus implying that neonatal gut and skin surfaces could be potential sources of CRKP which need special attention for infection control. Further, as the screening procedure involved both rectal and skin swabs, it was revealed that rectal colonization is more common than skin in these neonates.
Carbapenem-resistance in K. pneumoniae is mainly associated with blaKPC.13 However, molecular epidemiology in Asian countries is different, with predominance of metallo-β-lactamases (MBLs).22 All the CRKP isolates were positive for blaNDM in this study which is in concordance with the similar Vietnam based study.21 Wide dissemination of blaNDM and it being a MBL capable of hydrolyzing most β-lactam antibiotics, poses added challenges. This is because control of blaNDM and therapeutic options against infections caused by this gene is very limited.22 CRKP is of increasing clinical concern due to its transmission capacity. In this study, we described the multi-clonal dissemination of CRKP as detected by ERIC-PCR. Studies have suggested detection of several clones of CRKP in colonization, with dominance of clones like ST15 in Vietnam in their hospital in both admission and discharge positive CRKP isolates.21 The existence of multiple clones could suggest multiple sources of their acquisition rather than a common environmental source. Overlapping period of hospital stay in the NICU could increase the chances of transmission between the neonates of these CRKP isolates.23 It was interesting to note that similar clones were seen in rectal and skin colonization in the same neonate.
The study put forward several challenges in tackling these CRKP colonized neonates. Isolation and barrier nursing, though an idealistic approach, is not applicable in most of the resource limited setups in developing nations owing to shortage of service providers, high turn-over of patients and infrastructural constraints.24 Additionally, it was interesting to note that the mean age of the study population at admission was 3.73 ± 6.15 d comparable to the study which showed overall median age of neonates were 4 d.21 However, the mean age of admission of the CRKP colonized neonates in the present study was 1.40 ± 0.91, which was much less than the others. Consequently, it would be rational to speculate what could be the initial source of the CRKP from which the neonates got the colonization. In this regard, along with NICU environment, maternal gut has also been implicated as a possible reservoir of CRKP. Though maternal colonization could not be studied due to difficulties in logistics, a recent study on the impact of maternal colonization with Gram-negative bacteria with neonatal sepsis has not shown association between the two.25 Instead, NICU environment and nosocomial transmission had a role in sepsis. Surprisingly, in the present study, maternal complications were significantly associated with absence of CRKP colonization in the neonates and not otherwise. The authors of the present study have also reported existence of CRKP in the NICU environment in one of the previous studies, which could be the sources of colonization in these neonates.6
The study was not without limitations. The small sample size was one of the major limitation which was somewhat managed with multiple sampling (6 set of swabs per neonate). Secondly, weekly monitoring until discharge was not done which could have detected delayed infections and revealed the risk of infection with CRKP in the colonized neonates. However, the primary aim of the study was to detect the status of CRKP colonization in a CRKP endemic setup. Thirdly, sources of CRKP in these neonates were not investigated. Nonetheless, the strength of the study was that it added to the limited available data on neonatal CRKP colonization emphasizing the high burden and the urgent need of implementation of multifaceted infection control programmes in such CRKP endemic areas. Despite limited staffing, based on the study findings, we recommended routine CRKP screening in neonates for better care and understanding of their epidemiology.
The study revealed high burden of blaNDM carrying CRKP colonization in neonates with 20% CRKP colonization pressure at admission. This finding emphasizes the urgent need for the implementation of stringent infection control policies to prevent nosocomial transmission of these colonizers in infections in the neonates.
ACKNOWLEDGMENTS
The authors would like to thank Banaras Hindu University for providing the basic infrastructure required for the research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
KM, MS and PP collected the data, samples and carried out the experiments. SS and PR performed data tabulation and analysis. TB and AK planned and supervised the study. TB wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All data generated in this study are included in the manuscript.
ETHICS STATEMENT
The study was approved by the Institute Ethical Committee, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India, via letter No. Dean/2021/EC/2598.
INFORMED CONSENT
Informed consent was obtained from the neonates’ guardians before their inclusion in the study.
- Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11-20.
Crossref - Brooks B, Olm MR, Firek BA, et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6(1):112.
Crossref - Hartz LE, Bradshaw W, Brandon DH. Potential NICU Environmental Influences on the Neonate’s Microbiome: A Systematic Review. Adv Neonatal Care. 2015;15(5):324-35.
Crossref - Banerjee T, Bhattacharjee A, Upadhyay S, et al. Long-term outbreak of Klebsiella pneumoniae & third generation cephalosporin use in a neonatal intensive care unit in north India. Indian J Med Res. 2016;144(4):622-629.
- Banerjee T, Wangkheimayum J, Sharma S, Kumar A, Bhattacharjee A. Extensively Drug-Resistant Hypervirulent Klebsiella pneumoniae From a Series of Neonatal Sepsis in a Tertiary Care Hospital, India. Front Med. 2021;8:645955.
Crossref - Sharma S, Banerjee T, Kumar A, Yadav G, Basu S. Extensive outbreak of colistin resistant, carbapenemase (blaOXA-48, blaNDM) producing Klebsiella pneumoniae in a large tertiary care hospital, India. Antimicrob Resist Infect Control. 2022;11(1):1.
Crossref - Weterings V, Zhou K, Rossen JW, et al. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis. 2015;34(8):1647-1655.
Crossref - World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics 2017. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. Accessed 31st Oct 2023.
- WHO Country Office for India. Department of Biotechnology Government of India. Indian Priority pathogen list: to guide research, discovery, and development of new antibiotics in India. World Health Organization, Geneva, Switzerland. 2021:1-22. https://dbtindia.gov.in/sites/default/files/IPPL_final.pdf, Accessed 30th Nov 2023.
- Dias M, Saleem J. Surface colonization and subsequent development of infections with multi drug resistant organisms in a neonatal intensive care unit. Ann Clin Microbiol Antimicrob. 2019;18(1):12.
Crossref - Denkel LA, Schwab F, Kola A, et al. The mother as most important risk factor for colonization of very low birth weight (VLBW) infants with extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E). J Antimicrob Chemother. 2014;69(8):2230-2237.
Crossref - Loe MWC, Yeo KT. Early-life surface colonization with multi-drug resistant organisms in the neonatal intensive care unit. Int J Infect Dis. 2023;136:11-13.
Crossref - Akturk H, Sutcu M, Somer A, et al. Carbapenem-resistant Klebsiella pneumoniae colonization in pediatric and neonatal intensive care units: risk factors for progression to infection. Braz J Infect Dis. 2016;20(2):134-140.
Crossref - Ulu-Kilic A, Alp E, Percin D, et al. Risk factors for carbapenem resistant Klebsiella pneumoniae rectal colonization in pediatric units. J Infect Dev Ctries. 2014;8(10):1361-1364.
Crossref - Collee JG, Mackie TJ, McCartney JE. Mackie & McCartney Practical Medical Microbiology. 14th ed. Churchill Livingstone; 1996.
- Clinical Laboratory Standards Institute. Performance standard for antimicrobial susceptibility. M07-A11, 32nd edition. Introduction to tables 3B and 3C page no. 122. Test for carbapenems in Enterobacterales. Clinical and Laboratory Standards Institute, Wayne, PA. 2022.
- Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann Lab Med. 2012;32(5):359-361.
Crossref - Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-123.
Crossref - Fonseca EL, Ramos NV, Andrade BGN, et al. A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagn Microbiol Infect Dis. 2017;87(4):315-317.
Crossref - Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial enomes. Nucleic Acids Res. 1991;19(24):6823-6831.
Crossref - Berglund B, Hoang NTB, Lundberg L, et al. Clonal spread of carbapenem-resistant Klebsiella pneumoniae among patients at admission and discharge at a Vietnamese neonatal intensive care unit. Antimicrob Resist Infect Control. 2021;20;10(1):162.
Crossref - Veeraraghavan B, Pragasam AK, Bakthavatchalam YD, Anandan S, Swaminathan S, Sundaram B. Colistin-sparing approaches with newer antimicrobials to treat carbapenem-resistant organisms: Current evidence and future prospects. Indian J Med Microbiol. 2019;37(1):72-90.
Crossref - Mukherjee S, Mitra S, Dutta S, Basu S. Neonatal Sepsis: The Impact of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae. Front Med. 2021;11;8:634349.
Crossref - Pierce J, Apisarnthanarak A, Schellack N, et al. Global Antimicrobial Stewardship with a Focus on Low and Middle-Income Countries: A position statement for the international society for infectious disease. Int J Infect Dis. 2020;96:621-629.
Crossref - Robinson ML, Johnson J, Naik S, et al. Maternal Colonization Versus Nosocomial Transmission as the Source of Drug-Resistant Bloodstream Infection in an Indian Neonatal Intensive Care Unit: A Prospective Cohort Study. Clin Infect Dis. 2023;77(Suppl 1):S38-S45.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.