ISSN: 0973-7510
E-ISSN: 2581-690X
HCV transmission is commonly derived from blood transfusions. Some different aspects, such as location, infection prevalence, and genotype distribution, may affect the occurrence of HCV in blood donors. The blood donors have already been screened regularly for their anti-HCV serology, yet the test for HCV RNA has not been done yet. In this study, we aim to investigate the manifestation of HCV in Tuban by detecting HCV RNA from sera negative for HCV antibodies in blood donors. The blood donors from Tuban Red Cross Indonesia were recruited for a questionnaire interview and testing for HCV antibodies and HCV nucleic acids. Anti-HCV was serologically detected using ELISA. Nested PCR was used to amplify HCV-RNA in the NS5B and 5’UTR regions. The genotype or subtype of HCV is determined by direct sequencing followed by phylogenetic analysis. A total of 100 blood samples were collected. The HCV RNA positive rate was 6% in sera-negative anti-HCV blood samples. Furthermore, the genotyping resulted in 4 samples being dominantly HCV subtype 1c (66,67%); the other 2 samples were subtype 2a and type 1 (each counted as 1 individual, 16.67%, respectively). The serological test for HCV antibodies has been shown to be less sensitive than the nucleic acid amplification test. The detection of genotype 1c as a major HCV genotype circulating in the Tuban area may help to anticipate HCV transmissions and facilitate better medical treatment with respect to HCV carriers.
Hepatitis C Virus, Genotyping, Blood Donors
Infection with the hepatitis C virus (HCV) is a major issue worldwide, particularly in Asia. Around 180 million more instances of HCV are now prevalent globally, with a range of 64.4% in Asia to 74% in Australasia.1 Cirrhosis of the liver, hepatocellular carcinoma, and other liver-related morbidity and mortality cases are thought to be mostly caused by chronic HCV infection.
Transfusion-transmitted infections (TTIs) are the primary problem associated with blood transfusion. HCV is mainly transmitted through direct contact with the blood of an infected person, and 90% of HCV transmission frequently occurs through blood transfusion. Recently, HCV transmission from blood transfusions decreased in number and only accounted for 4% of total HCV cases. Meanwhile, 50% of HCV cases are transmitted through the misuse of intravenous drugs.2,3 Other methods of transmission include tattoos, scars, and hemodialysis (this is possible because the infected people used non-sterile equipment). The chances of contracting HCV are also greater in transsexuals, people who change sex partners frequently, and children whose mothers have the virus. Only when the concentration of HCV RNA is large enough can HCV transmission occur under such conditions.3,4
HCV has 50 subtypes that are represented by seven primary genotypes, designated 1 through 7, and various regional variations in the genotype distribution.5,6 The issues emerge from how difficult it is to create a single, all-encompassing HCV vaccination. Given that it is a pricey test, HCV genotyping is not particularly frequent in Indonesia. Although different genotypes don’t produce different symptoms, some medications can lead to difficulties when this virus is exposed to them.5-7
Historically, Tuban served as a significant port for the East Javan economy, bringing quickly urban activity from Indonesia or other nearby nations. Given that this metropolis has incorporated many foreign influences, it is essential to do community-based HCV screening, beginning with blood donors. It was necessary to carry out this in order to look at how the Tuban people contracted HCV. As a result, the objective of our present research is to identify the HCV genotypes in HCV-RNA positive blood donors in order to prevent TTI. Expectedly, our findings can be utilized to monitor the blood supply’s safety, which could lower the number of cases of HCV transmission by blood transfusion.
Distribution and sample collection
The participants were selected from a blood transfusion center in Tuban, East Java, Indonesia, and 100 sera samples from blood donors were collected with etichal clearence. The participants had signed informed consent. The study protocol was performed according to the Helsinki declaration and approved by local ethics committee Medical Faculty, University of Airlangga, Surabaya, East Java, Indonesia. Informed written consent was obtained from people who perform blood donors at Indonesian Red Cross Foundation in Tuban, East Java. Sample collection 2 July 2015 to 30 July 2015. Inklusi criteria patient antisera negative.
HCV antibody was detected by enzyme-linked immunosorbent assay (ELISA), followed by the detection of HCV RNA by nested PCR. For anti-HCV detection, all procedures are based on ELISA’s kit instructions using the Foresight® EIA test kit for HCV, cat. no. 1231-1031 (ACON, California, USA). Extraction of HCV RNA from anti-HCV negative sera samples using the Qiagen RNA extraction kit (Qiagen GmbH, Hilden, Germany)
RNA extraction of HCV and reverse transcription
HCV cDNA synthesis was done by changing HCV RNA to HCV cDNA using the reverse transcriptase enzyme and a random hexamer primer. and reverse transcriptase by RT-PCR kit (Toyobo Inc., Osaka, Japan) as described in our previous publication.8
Nucleic acid testing using nested PCR and Direct nucleotide sequencing
For HCV RNA amplification, nested PCR was used for targeting the NS5B region, and if the result was negative, it then continued to nested PCR for targeting the 5UTR of HCV. The primers used were from three different references. The first primer is used to amplify the NS5B region.8,9 Primers NS5B-F1 (nt 7999–8020), 5-CAATWSMMACBACCATCATGGC-3, and NS5B-R1 (nt 8805–8825), 5-CAGGARTTRACTGGAGTGTG-3, are used in the first round of PCR. Furthermore, for the second-round PCR primers NS5B-E2 (nt 8159-8181), 5 ATGGGHHSBKCMTAYGGATTCC-3, and NS5B-R2 (nt 8611-8630), 5’-CATAGCNTCCGTGAANGCTC-3’ (H77 strain references, GeneBank accession number AF00906) were used.
Following the negative results from the first PCR targeting the NS5B region, another nested PCR was done using a second set of primers (also for region NS5B). For the second nested PCR, the primer sets used for the first round were as follows: F-166 5-TGGGGATCCCGTATGATACCCGCTGCTTTGA-3′ (8230-8260, +) and R-167R 5′-GGCGGAATTCCTGGTCATAGCCTCCGTGAA-3′ (8601-8630, -). 10,11 For second-round PCR, there were several sets of primers listed below (odd numbers are forward primers, even numbers are reverse primers) (Apichartpiyakul, 1994):
HC23 5 ‘-TTTGACTCAACCGTCACTGA-3’ (8256-8275, +),
HC24 5 ‘-CTCAGGCTCGCCGCATCCTC-3′ (8577-8596, -),
HC26 5′ -CTCAGGTTCCGCTCGTCCTC-3′ (8577-8596, -),
HC15 5′-ACTGTCACTGAACAGGACAT-3’ (8265-8284, +)
HC16 5 ‘ -GCTCTATCCTCATCGACGCC-3′ (8568-8587, -)
HC28 5′ -CACGAGCATGGTGCAGT CCCGGAGC-3′ (8507-8531, -)
HC32 5′ -AGGTAGCACGTCAGCGTG TTTCC-3′ (8454-8476, -)
HC34 5′ -TAGCACGTCATGGTGTTTCCCAT-3’ (8451-8473, -) as previously mentioned in our publication.12
For the remaining samples negative for NS5B region amplification, another round of nested PCR was used by targeting the 5’UTR region. The first round of nested PCR for the 5’UTR region was performed using UTR1 (sense; 5’-CCGGGAGAGCCATAGTGGTC-3’) and UTR2 (antisense; 5’-AGTACCACAAGGCCTTTCGC-3’). The second-round PCR was performed using UTR3 (sense; 5’-TGGTCTGCGGAACCGGTGAG-3’) and UTR4 (antisense; 59-ACCCAACACTACTCGGCTAG-39) as an inner primer set.13
PCR amplification was done utilizing “GeneAmp PCR System 2400 (Perkin Elmer)”, each PCR consist of 40 cycles. The cycles begin with hot start at 94°C for 5 minutes. Cycles consist of denaturation at 94°C for 60 seconds, annealing (temperature based on the primer used) for 60 seconds and elongation at 72°C for 75 seconds.
PCR product visualized using electrophoresis using TBE buffer 1X and using agarose gel 2% that contained ethidium bromide. Visualization had done using Ultra Violet (UV) transilluminator.
Purification of PCR products was done with the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). Following the purification steps, the HCV DNA samples were then labeled with reagent for Big Dye Terminator Kit V.1.1 before finally being sequenced using the Sequencer Machine ABI PRISM-310 (Applied Biosystems, Inc.).
Molecular phylogenetic analysis
A phylogenetic tree was created using the nucleotide sequences of HCV that were obtained from the sequencing data (just for NS5B data). In order to identify the HCV genotypes and subtypes (using reference sequences acquired from GeneBank), the alignment analysis was also carried out (just for 5’UTR data). GeneTYX-Win 10 has been used for all nucleotide analysis. At Universitas Airlangga’s Institute of Tropical Disease, sequencing and genotyping analysis were conducted.
HCV RNA identification and Phylogenetic tree
All samples from Tuban blood donors were serologically checked for anti-HCV, and then nested PCR was used to check for HCV RNA. Only six (6%) of the 100 samples collected tested positive for HCV RNA. 83.33% of those with HCV RNA were men (5/6) and 15.67% were women (1/5), according to the results of the analysis. According to this finding, men were more likely than women to have HCV infection when donating blood in Tuban.
Men who had a negative antibody response to a particular virus, in this case called HVC, as many as 85 people, while women who had a negative antibody response to the virus were only 15 people. The results of the PCR examination showed that of 85 illustrations of men who had a negative antibody response to HVC, there were 5 people whose PCR test results were positive. On the other hand, from 15 samples of women with negative antibody responses to HVC, only 1 person had a positive PCR test result.
From the results of this study, it can be concluded that the number of men infected with HVC is more than the number of women. When related to the COVID-19 virus, people with HVC have a comorbid opportunity (other health conditions that coincide) so they are more susceptible to being infected with the COVID-19 virus. That is, people infected with HVC may have a weak immune system or have health conditions that make them more susceptible to inflammation of the COVID-19 virus.
However, keep in mind that this data is only sourced from research data researchers do not include bonus data such as age, medical history, or other risk aspects. Further research may be needed to master the link between HVC inflammation, susceptibility to COVID-19, and other factors that could influence these results.
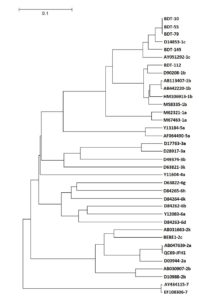
From 100 samples, 94 remain negative for HCV RNA after being tested by nested PCR. In the six samples that were positive for HCV RNA, 83.33% (5/6) were positive in the NS5B region. Subsequently, these samples were sequenced and visualized in a phylogenetic tree for HCV genotyping analysis. A phylogenetic tree is depicted in Figure. The sequencing results revealed that 20% (1/5) of the samples were identified as subtype 1b (sample code: BDT-112) and 80% (4/5) belong to subtype 1c in the NS5B region (sample codes: BDT-10, BDT-55, BDT-79, and BDT145) (Figure). The remaining samples were HCV RNA-positive in the 5’UTR region, and genotyping analysis revealed they were categorized as HCV type 1 (no subtype).
In the first year of this investigation, blood samples from Tuban blood donors were gathered and examined for anti-HCV serology. Three major groupings appeared from the 100 samples of these samples that tested negative for the anti-HCV virus. A total of 50 samples had anti-HCV negative levels that were deemed high (near cut-off values), 25 samples had intermediate anti-HCV negative levels (under cut-off values), and 25 samples had extremely low anti-HCV negative levels. All 100 samples of anti-HCV negative sera were subjected to PCR in the second year of this trial, and six samples (or 6%) were found to be positive for HCV RNA. The samples from all three categories high, middle, and low were HCV-RNA positive.
The detection of positive HCV RNA from anti-HCV negative sera samples indicated that HCV carriers could be identified when the individuals were in the “window period”, which means HCV RNA is already being produced in the blood yet HCV antibody levels are too low to be measured by a commercial ELISA kit. Based on our results, the prevalence of HCV infection during this “window period” was counted for 6% of the blood donors in Tuban. This number is possibly categorized as high considering the Tuban population, and it needs immediate attention and further action. According to this predicted number, the virus can cause infection for the donor itself and a higher risk of HCV transmission to the blood recipients (transfusion-transmitted infections). Akbaylar et al.14 found that HCV RNA was detected in the sera of 53 out of 68 (77,9%) patients who were anti-HCV negative.
From 100 anti-HCV-negative sera, six HCV RNA-positive samples have been successfully found. Six samples were examined, and four (66.67%) were found to be subtype 1c in the NS5B region of HCV, while one sample (16.67%) belonged to subtype 1b. In the 5’UTR region, the remaining sample has genotype 1 HCV (no subtype) shown in Table.
Table :
HCV Genotypes of anti-HCV negative sera from blood donors
No. |
Sex |
Primers |
Genotype |
Total |
|---|---|---|---|---|
1. |
Man |
NS5B |
Subtype 1b |
1 |
2. |
Man |
NSB5 |
Subtype 1c |
4 |
3 |
Woman |
UTR |
Type 1 |
1 |
Total |
6 |
Furthermore, differences in geographic location can cause differences in genotype distribution, so further research is needed to determine differences in genotype distribution in each region in Indonesia. Viral genotype is also used to predict response to antiviral drugs.15,16 For example, administering sovosbuvir in combination with ribavirin is not recommended for genotypes (1, 4, 5, and 6). The high frequency of genotype 3 in a country is a problem in itself because genotype 3 tends to be difficult to treat and has a poor prognosis.16
Since traditional serological testing for HCV antibodies have been shown to be less sensitive than the nucleic acid amplification test, HCV infection continues to be a significant public health issue in Tuban. HCV genotype 1 (subtype 1c) was found to be the most prevalent HCV genotype in Tuban among blood donors. The discovery of genotype 1c as a prevalent HCV genotype in the Tuban region may aid in the prediction of HCV transmissions and improve medical care for HCV carriers.
ACKNOWLEDGMENTS
The authors would like to thank the Ministry of Research, Technology, and Higher Education of Indonesia for the financial support of this study, Center of Blood Transfusion – Tuban for providing sample collection, and Institute of Tropical Disease (ITD) – Universitas Airlangga for facilitating experimental works required for this study.
CONFLICT OF INTEREST
The authors declared that there was no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the Directorate of Research and Community Service in 2016 from Ministry of Research, Technology, and Higher Education of Indonesia.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Petruzziello E, Margliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22(34):7824-7840.
Crossref - Lavanchy D. Evolving Epidemiology of Hepatitis C Virus. Clin Mcrobiol Infect. 2011;17(2):107-115.
Crossref - Friedman LS. Liver, Biliary Tract, & Pancreas Disorders. Current Medical Diagnosis & Treatment. Fifty- Second Edition. Edited by: Papadakis MA, McPhee SJA, Rabow MW. New York, McGraw-Hill Education 2013; 662-682.
- Allison RD, Camtilena CC, Koziol D, et al. A 25-Year Study of the Clinical and Histologic Outcomes of Hepatitis C Virus Infection and Its Modes of Transmission in a Cohort of Initially Asymptomatic Blood Donors. J Infect Dis. 2012;206(5):654-661.
Crossref - Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of Hepatitis C Virus genotypes. Hepatology. 2005;42(4):962-973.
Crossref - Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’untranslated region sequences. J Clin Microbiol. 2007;45(4):1102-1112.
Crossref - Juniastuti, Utsumi T, Nasronudin, et al. High rate of seronegative HCV infection in HIV-positive patients. Biomedical Rep. 2014;2(1):79-84.
Crossref - Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, et al. Geographic distribution of hepatitis C virus genotype 6 subtypes in Thailand. J Med Virol. 2010;82(2):257-262.
Crossref - Anggorowati N, Yano Y, Heriyanto DS, et al. Clinical and virological characteristics of Hepatitis B or C Virus co-Infection with HIV in Indonesian patients. J Med Virol. 2012;84(6):857-865.
Crossref - Mori S, Kato N, Yagyu A, et al. A new type of hepatitis C virus in patients in Thailand. Biochem Biophys Res Commun. 1992;183(1):334-342.
Crossref - Apichartpiyakul C, Chittivudikarn C, Miyajima H, Homma M, Hotta H. Analysis of hepatitis C virus isolates among healthy blood donors and drug addicts in Chiang Mai, Thailand. J Clin Microbiol. 1994;32(9):2276-2279.
Crossref - Nurtjahyani SD, Amin R, Hndajani R. Detection of hepatitis c virus RNA in blood donors with nested PCR technique. Biomedical Engineering. 2016;2(2):1-5.
- Doi H, Apichartpiyakul C, Ohba KI, Mizokami M, Hotta H. Hepatitis C Virus (HCV) subtype prevalence in Chaiang Mai Thailand, and identification of novel subtype of HCV mayor type 6. J Clin Microbiol. 1996;34(3):569-574.
Crossref - Akbaylar HA , Abacioglu H , Tanhurt E, et al. Prevalence and Genotyping of hepatitis C virus RNA in Turkish patient with chronic non A non B liver disease. Turk J Gastroenterol. 1998;9(3):208-212.
- Nurtjahyani SD, Probojati RT, Ansori ANM, Amin M, Handajani R. Haplotype Network Analysis and Phylogenetic Tree Construction of Hepatitis C Virus (HCV) Isolated from Tuban, Indonesia. Research J. Pharm. and Tech. 2021;14(8):4231-4235.
Crossref - Utama A, Tania NP, Dhenni R, et al. Genotype diversity of hepatitis C virus (HCV) in HCV-associated liver disease patients in Indonesia. Liver Int. 2010; 30(8): 1152-1160.
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.