ISSN: 0973-7510
E-ISSN: 2581-690X
Microbes proved to be the significant biotic factors that influence the health of humans. Gut microbiota remains an emerging field for understanding different aspects of microbiology, immunology, computational biology and food and nutrient supplementation studies. The human microbiome project provides a thread in the path of microbe association with humans. This review will discuss how their study was taken last year on human microbiome discovery for human health. Thus, the microbiome could be deliberated as target for treating various disorders. Despite some limitations, interventions in this field of study appear encouraging for emerging a preventive therapy by restoring microbiome functionality or as an adjuvant in specific immunotherapy. Manipulation of the gut microbiota in various disorders is assessed by examining the current most relevant evidence concerning to antibiotics, probiotics, prebiotics, polyphenols, and fecal microbiota transplantation. This review discusses the impact of gut microbiota on health and their manifestation by focusing on vital mechanisms.
Microbiota, Disease, Nutrient, Intestine, Dysbiosis
A microbiome is defined as the set of microorganisms living on or inside living organisms. This microbiome includes bacteria, fungi, viruses, and protozoa that consist of 150 time’s greater genetic material than that of the host.1 The well-studied niche microflora is inside the human gut, where about tens of trillions of microbes live symbiotically. Phyla, Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria are, more prominent in healthy adults.2 When the species of different class of microbes are deregulated, it contributes to several complication related to the health. In human body, colon contains the highest microbial diversity that is present on earth. In human gut Bifidobacterium, Bacteroides, Prevotella, Fusobacterium, Eubacterium, Peptococcus, Peptostreptococcus Ruminococcus, class of microbes abundant. Escherichia and Lactobacillus. Bacteroides alone constitute about 30% of all microbes in GI tract.3 Innate and adaptive immune systems of host also contributed by microbiome.4,5 Dysboisis of microbiome cause inflammation and autoimmune diseases.6-8 Intestinal dysbiosis deregulates metabolic activities which are highly associated with host factors like age, diet, and environmental conditions.9 Accompanying mechanistic exploration toward host microbe interaction, in 2007 human microbiome project (hmpdacc.org) started focused on identifying and characterizing human microbial flora.10 Hence intestinal microbe can be major target for health improvement.11
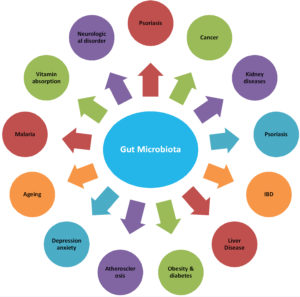
Microbiome specific disease diagnosis, monitoring and treatment may revolutionize much incurable drug dependent disease worth anticipating. Microbiome technologies are expected to play a crucial role in the development of new drug discovery platforms and companion diagnostics in the precision medicine era (Figure 1).
Common Enterotype in Gut Ecology
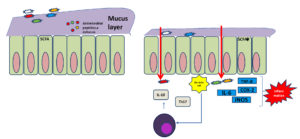
Human gut microbiome has been classified into three distinct enterotypes (Bacteroides, Prevotella, and Ruminococcus). Gut microbial enterotypes are associated with specific fooding habits and drug intake patterns. Prevotella– are enriched in people consume carbohydrate diet. While high protein and fat diet intake dominate Bacteroides– population. Ruminococcus-dominant enterotype is associated with grain Lovers. 16S rRNA gene of human intestinal tract microflora explores diversity measurement.12,13 Metagenomics studies also characterized functional microbiome among American and Japanese individuals. The intestinal flora includes Lactobacillus, Bifidiobacteria, Peptostreptococci Propionibacteria and Enterococci.14,15 Primary function of these microorganisms is to live in commensal and protect gut after secreting antibiotic substance that inhibit the growth of harmful microorganism and maintain pH that is essential for intestinal wall to perform protective barrier, and provide an environment where microbe cannot colonize.16-20 Opportunistic flora, of intestinal include Clostridia, Bacteroides, Staphylococci, Streptococci, Bacilli, Yeasts, Peptococci Enterobacteria, Fusobacteria, Eubacteria, Catenobacteria and others. The flora which enters the body through food and drink constitutes the transitional flora.21 Dysbioisis of microbiome cause inflammation and autoimmune diseases,6-8 promoted by loss of intestinal barrier function, against which an immune cascade get activated (Figure 2).
Figure 2. Presentation of eubiosis and dysbiosis difference in intestine, microbes maintain gut membrane stability with production of certain metabolite (i.e short chain fatty acid, butyrate) and prevent microbial expansion. In dysbiosis specific microbe’s overgrow and metabolites changes in absence of specific species increase production of proinflammatory cytokines such as IL-6, IL-17, TNF-α and cause inflammation
Psoriasis and Psoriatic Arthritis
Psoriasis is an autoimmune disease in which skin cells grow faster than normal and formed a bumpy red patch covered with white scales.22 Several studies on surface or skin microbiota confirmed the elevated Staphylococcus and Streptococcus genus.23 In psoriatic patient bacterial DNA has been isolated both locally and systemically, and altogether evidence a central role of bacteria in psoriatic disease.24
Prolonged psoriasis illness cause damage of tissues and bone which resulted in psoriatic arthritis.22 Gut microbiota study of psoriatic patient clear the abundance of Prevotella copri.23 Decrease Bacteroides genus and an increase in the numbers of Faecalibacterium, Akkermansia, and Ruminocuccus genera is also a characteristic findings in the gut microbiome of patients with psoriasis.11 African cluster study found that prevotella species, predominantly contributes to the joint inflammation in the intestine. Auto reactive T cells turn out to be highly reactive to auto antigens, including arthritis specific antigens, via activation of innate immunity.24,25 These T cells populations can then exacerbate joint inflammation. Presence of bacterial DNA in psoriatic patient plasma confirmed.26 This increased bacterial load in blood arose from intestinal barrier disruption, along with their metabolite access to blood stream and skin homeostasis impairment.27,28 These findings established a correlation between the gut microbiome and skin homeostasis. Psoriatic skin had less Propionibacterium and Corynebacterium had less Ruminococcaceae and Akkermansia but more Bacteroides and Faecalibacterium.28,29 Intake of oral probiotic of Bifidobacterium infantis35624 reduced the plasma levels of TNF-a as compare to control.30 Oral administration of L. salivarius LA307 and L. rhamnosus LA305 showed reduction of eczema and inflammation marker.31 In imiquimod-induced psoriasis mouse model L. pentosus GMNL-77 and Lactobacillus sporogenes administration (as a probiotic) were able to suppress TNF-a, IL-6, and proinflammatory cytokines in the IL-23/IL-17 cytokine.32
Inflammatory Bowel Disease (IBD) and Colitis
Chronic Inflammation of small intestine, colon and Crohn’s disease (CD) and ulcerative colitis (UC) are called inflammatory bowel disease.33 Gut dysbiosis, and dysfunctional immune responses cause intestinal tissue damage by activated Th1 and Th17.34 Mucosal injury results in invasion of microbial antigens, TLR ligands that perpetuate the immune responses.35 NOD2 gene present in human encodes intracellular pattern recognition receptor that recognizes bacterial peptidoglycan and stimulates immune system.36 A report showed that NOD2 gene knockout in mice tends to decrease IL-10 and increased susceptibility to colitis, in human subjects with NOD2 mutations.37 Increases in the Gammaproteobacteria and decreased abundances of bacterial taxa of Firmicutes and Bacteroides.38,39 Most common microbes present in inflammatory bowel disease patient is Rhodotorula, which is capable of inducing intestinal inflammation.40 transfer of these bacteria induces inflammation in healthy mice. In CD bacteria which produce short chain fatty acid Phascolarctobacterium and Roseburia are also reduced while Leuconostocaceae is depleted in UC along with enrichment of Escherichia species and a depletion of Faecalibacterium species.41,42 Bacteroides vulgatus bacteria alter mucosal membrane function and turn on inflammatory genes.43,44 Nonpathogenic strain of E. coli used as prebiotic has been shown effective in patients of ulcerative colitis.45 B fragilis prevents colitis in mice by interacting with CD4 Foxp3 regulatory T cells which produce IL-10.46
Therefore bacteria and fungi can penetrate mucosal barrier and activate TLRs, CARD9 and Decting-1 activation resulted in more severe phenotype in lamina propria.47,48 When CARD9 knockout in mice, they got susceptible to colitis. This susceptibility was directly associated with tryptophan metabolism correlated to impaired microbial population.49
Carbohydrate fermentation products, propionic acid, acetic acid and butyric acids which play important role in activation and regulation of multiple pathways specific to colon.50 Among all metabolites butyrate significantly involved in colonocytes metabolism.51 This imparts anti-inflammatory action by inhibiting IL-12 and TNF-α cytokines and up regulating IL-10 in human monocyte.52 Expansion of T-Cell to mitigate intestine inflammation induces by Bacteroides and Clostridium species.53 Previous studies on IBD suggest that combination probiotic of Bifidobacterium breve, Bifidobacterium infantis, B. longum, L. acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Streptococcus thermophiles, and Lactobacillus bulgaricus have potential to reverse ulcerative colitis.54 Mesalamine, an anti-inflammatory drug decreases the Escherichia/Shigella in IBD and attenuates intestinal inflammation.55,56
Liver Diseases
Liver is the major vital organ that detoxifies the body and support function of other organs. Failure of liver function can leads to serious health complication. Relation between liver diseases and gut microbiota has been well established.57
Imbalance in pathobiont and beneficial bacteria is associated with deleterious effects on host and plays a critical role in the development of alcoholic liver disease,58 nonalcoholic fatty liver disease,59 nonalcoholic steatohepatitis,60 acute (toxic) liver injury, fibrosis/cirrhosis, hepatic tumors,61 autoimmune hepatitis and autoimmune cholangiopathies.62 These disorders arise from translocation of bacterial metabolites by intestine to liver.63 These endotoxin metabolites received via toll-like receptors and cause damage to liver associated with higher levels of serum TNF-α, IL-6, and adipokines 2.64 Moreover, bile acid dysregulation linked to dysbiosis, hepatic lipid accumulation increases insulin resistance, and inflammatory signaling.65
When infection of Streptococcus pneumonia increase in liver, a protein that is reactive to the C-polysaccharide of bacteria called as C-reactive protein take part in antigen removal.66,67 Concentration of CRP in blood directly proportional to inflammation in response to infection, trauma, and tissue infection, Hence, the CRP is monitor in various inflammatory conditions.68 Higher faecal proportions of Bifidobacteria, enterobacteria and streptococci associated with alcoholic hepatitis.69
Thus, intestinal microbiota is center for progression of alcoholic liver disease. Conversely, Proteobacteria, Bacteroidetes, and Enterobacteriaceae were found to be increased in NASH patient as compare to healthy controls, while Firmicutes declined.70 Hence Bacterial translocation cause systemic infections in liver of Proteus and Enterobacter which have ability to bypass the intestinal barrier and associated with higher incidence of bacteremia. In liver cirrhosis, population of Bacteroidetes is significantly reduced, while Proteobacteria and Fusobacteria increased.71 Mixture of Bifidobacteria, Lactobacilli, and Streptococcus thermophiles probiotic was able to reduced high-fat diet (HFD)-induced steatosis, inflammation, and insulin resistance in a rodent model.72
Obesity and Type 2 Diabetes Mellitus
Obesity and diabetes are the third most common disease in aged people after cardiovascular disease and cancer.73,74 It’s a debate where obesity cause type 2 diabetes, high blood pressure and heart disease or because these illness person became obese.75 Obese people are always on risk of developing type 2 diabetes. In obese persons, fat tissues cells have to process more nutrients than their regular capacity.76
In metabolite study of plasma and stool samples 19 metabolites associated with BCAA (branched-chain and aromatic amino acids) linked to high BMI and obesity development.77 Diet consisting saturated fat, associated with increased inflammation with abundant white adipose tissue (WAT) and metabolic disease, on the other hand polyunsaturated rich diet counteract inflammation and promote healthy lean phenotype due to changed metabolism.78 Fat induced WAT accumulation occurred as a result of inflammation, mediated through gut microbial activation of TLR4 which serve as receptor for microbe-associated molecular pattern (MAMP) receptors, and further activate proinflammatory response through nuclear factor (NF)-kB pathway, producing antibacterial products, cytokines and chemokines.79 Studies performed in different diabetic group shown lower intestinal Roseburia and Bacteroidetes. Abundant Faecalibacterium prausnitzii (both butyrate producing bacteria), and other strains Lactobacillus gasseri, Streptococcus mutans, Firmicutes and Clostridiales members also contributes to disease.80
In a experiment where germ free mice inoculated with obese (ob/ob) or lean (+/+) microflora. Mice that received obese microbes extracted more calories from their food and increase in total body fat than in mice colonized with lean microbiota. In one study author insert the lean phenotype microbiota in obese phenotype, and find improve insulin sensitivity.81 Resistance of insulin reported as a increase in branched chain amino acid which are associated with gut microflora.82 Changes in metabolites related to obesity were directly linked to four different bacteria (Ruminococcus, Blautia, Dorea and in the Lachnospiraceae family, and SHA98.83 Supplementation of A. muciniphila reduced inflammation reduces LPS level and improved glucose tolerance.84,85 Fermented probiotic fiber has potential to protect against metabolic syndrome86 consequence of reduced intestinal permeability.
Atherosclerosis
Atherosclerosis is the deposition of fat molecule on artery wall, Plaque of fat and other minerals reduce the diameter of artery for the flow of oxygenated blood. There are some factor like high LDL, obesity, smoking and consumption of unhealthy diet results of this condition along with family history. Western diet comprises diet reach in choline and carnitine have increased risk of cardiovascular disease.87,88 Gut microflora convert these compound into trimethylamine (TMA), which is further interconvert into trimethylamine- N-oxide in the liver and trigger heart problems.89-91 This conversion of TMA catalyzed by microbial TMA lyase. Atherosclerosis development triggered by TMA through up regulation of the CD36 of macrophages and promotes uptake of cholesterol and formation of foam cells.92,93 Reduced expression of cytochrome P450 7A1 and 27A1 which metabolize drug and other molecule in liver, and promotes vascular inflammation by activating nucleic acid factor-κB signaling pathway, monocytes through mitogen-activated kinase.94-96 Inhibits cholesterol reverse transcription.97,98
Gut microbiome composition in atherosclerotic patients observed to be more inflammatory.99 Elevated Escherichia coli, Klebsiella spp., Enterobacter aerogenes, Streptococcus spp., Lactobacillus salivarius, Solobacterium moorei, and Atopobium parvulum. Depleted abundance of gut species such of Bacteroides spp., Prevotella copri, and Alistipes shahi100,101 has been recorded in patient. Administrative study of Lactobacillus plantarum as probiotic reduce blood cholesterol and therefore restrain the formulation of atherosclerotic plaques in hyper-cholesterol patients.102 When A. muciniphila supplemented in atherosclerosis prone mice, it protected atherosclerosis development while mice were feed on Western diet.103 Indeed, mice harboring high choline-metabolizing bacteria are susceptible to diet-induced metabolic disease.104 Secondary metabolite of polyphenol class like Gallic Acid a-Asarone, Equol, Urolithins and Quercetin-3-glucuronide function in protection of cardiovascular system.105,106
Microbial metabolism modulation through diet intervention or direct supplementation provides effective strategy for prevention of cardiovascular diseases.
Ageing
Aging is a consistent decline in vital activities that is associated with inflammation. Age dependent changes are associated with oxidative/ genotoxic stress and change in gut microbiota composition by the time.107,108 Bifidobacteria by the span of life decline, whereas Clostridium perfringens, Lactobacilli, and Enterococci increase with age.109 Composition change of gut microbiota directly correlated with intestinal inflammatory state.110 Current studies on microbiome and aging associated changes evidenced the role of certain population which regulates pro-inflammatory and anti-inflammatory networks in the gut microbiota.111,112 Inflammatory markers such as LPS-binding proteins increase in aged healthy person.113,114 Hence low grade inflammatory status facilitates by LPS.115
Malaria
Malaria is a mosquito transmitted disease affect 219 million population yearly, Plasmodium species is causative agent. Different mice studies showed that mice devoid of commensals bacteria are more susceptible to Shigella flexneri, Bacillus anthracis, and Leishmania infection.116,117 Moreover, gut microbiota can diminish intestinal immunity and increase susceptibility toward enteric pathogens such as Citrobacter rodentium and Campylobacter jejuni.118,119 Providing the importance of the microbiota in regulating immune system function homeostasis.
Mice study Demonstrated E. coli expressing a-galactosyl, host were protected from effective Plasmodium liver-stage infection.120 Influence of the microbiota on the progression of malaria clinical outcome infections has only now started to emerge.
Absorption of Vitamin B12
Vitamin B-12 or cyanocobalamin is essential co-factor for iron metabolism as well as nucleotide synthesis and neuronal system function. Deficiency could introduce tiredness, anemia, constipation, weight loss, memory problems, and neurological issues.121 Major problem arise either deficiency in food or malabsorption due to lack of bacterial population helps in interconversion in intestine.122 Cobalamin deficiency cause less cell division, neuropathy, nervous system disease, and pernicious anemia.123 B12 deficiency is prevalent in vegetarian diet people as plants are not a source. Concentrations of cobalamin can be detected in fermented food.124 An intrinsic glycoprotein factor secreted by gastric mucosa serves as important absorption factor. Cobalamin deficient patient’s microbiota showed dramatical changes after cyanocobalamin and methylcobalamin supplementation.125 Methylcobalamin alleviate butyrate producing bacteria which may alleviate intestinal inflammation. Previous studies confirmed that vitamin B12 deficiency is also contributes to inflammatory bowel disease.126
Neurological Disorders
Gut is also called second brain of human, so disorder in neurological system state the gut health disturbance that is consequence of microbe ecosystem misbalance.127 Neurological complication aroused due to failure of gut membrane function that allow other microbes and their metabolites to blood stream as a loss of permeability,128 Which may disrupt the synthesis of SCFA (short chain fatty acid) served as vitamin and hormone cofactors,129-131 and generate molecules that reduce inflammation,132 and play a function to boost immune activities. Medium chain fatty acid, alpha lipoic acid which is the activator of the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) which response in antioxidant element signaling pathway.133,134 Increased inflammation of gut linked to neurodegenerative diseases.135-137 Gut microbiota alteration have been directly linked with mental comorbidities like depression, multiple sclerosis and Parkinson disease. Structural, biochemical or electrical abnormalities in the brain, spinal cord cause physical impairment such as neuromyelitis optica spectrum disorders, multiple sclerosis, Parkinson disease, Alzheimer disease, Huntington disease, and amyotrophic lateral sclerosis.138-142 Enrichment of Archaea (genus Methanobrevibacter) was relative to controls as well as depletion of members from the Firmicutes (eg, Clostridium genera) and Bacteroidetes phyla reported. Consumption of antioxidant throughout life can also impede neurological symptoms along with enhancement of immune system. Neurological diseases are mainly can be cured through cleaning of colon. Certain Ayurveda remedies also showed very tremendous result after a panchkarma procedure of “Basti” (rejuvenation of large intestine).143
Depression and Anxiety
Role of gut microbiome has been implicated in various disorders, with particularly strong evidence for its role in depression and neuropsychiatric disorder.144 Post gut microbiota transfer of depression patient healthy mice showed elevated level of tumor necrosis factor alpha (TNF-α).
Interleukin-6 (IL-6), interleukin-8 (IL-8), along with C-reactive protein (CRP) and higher kynurenine/tryptophan ratio. Total blood cortisol was also increased which is reflecting stress environment in body. Patient data showed
Profound difference in Prevotellaceae family and Prevotella genus. Short chain fatty acid, plasma lipopolysaccharide binding protein did not directly correlate. Although, Alistipes, Faecalibacterium, and Ruminococcus were correlated with a detectable level of isovaleric acid.145 Response of gut microbes is gender biased; Lactobacillus rhamnosus did not improve stress in male.146 Protiobic species Lactobacillus helveticus and B. longum are able to reduce depression in patients with major depressive disorder (MDD).147 These results are contradictory with negligible effect on clinically depressed patients.148 Although, animal model able to produce the human relevant data, but investigation of human samples to establish a proper relationship investigating mechanism behind it is needed. Encouraging results of probiotics.
Cancer
Cancer is a leading cause of death worldwide. In 2018, cancer accounted for 9.6 million deaths.149 Role of numerous gut populating bacteria, have been recognized as protectant against the tumors genesis.150 Lactobacillus rhamnosus GG (LGG).
Is the most studied probiotic for cancer intervention studies.151 In colon cancer patient, change in bacterial community has been reported with abundance of pathogenic bacteria.152 Lactobacillus and Bifidobacteria prevent tumor development by MyD88 suppression, which play essential role in developing carcinomas.153,154 Lactobacillus johnsonii modulate immune system in colorectal patient by Enterobacters modulation.155 In an experiment with colonized Helicobacter hepaticus mice exhibit colonic Th17 inflammatory molecule also have a beneficial role in human ovarian cancer.156,157 murine melanoma, pancreatic and colon cancer.158,159 Helicobacter pylori is known for alter stomach pH and acid reflux, which could protect against Barrett’s esophagus and esophageal cancer.160,161
Kidney Disease
Various combination of different type of disorders like high blood pressure, diabetes, urinary tract obstruction, kidney stone, enlarged prostate are also part of pyelonephritis. In kidney disease lower intestinal microflora has been found to be altered mostly associated with decreased Prevotellaceae and Lactobacillaceae families.162 Analysis of fecal microbiota showed overgrowth of aerobic bacteria, such as Enterococci and Enterobacteria species, was 100 times greater in hemodialysis patients along with Lower numbers of Bifidobacteria and higher Clostridium perfringens.163 In Chronic Kidney Disease urea secrete into the gastrointestinal tract where hydrolysis of urea takes place by urease which is expressed by some gut microbes. Ammonia produced in gut which can affect the growth of commensal bacteria.164 Mild consumption of fiber diet, use of antibiotic, intestinal wall edema, and oral iron intake, metabolic acidosis also cause dysbiosis.165 Nephroprotective metabolite such as vitamin K and butyrate lowered due to microbes producing (e.g., short chain fatty acids), such as Lactobacillus.166 Gut bacteria Clostridium and Bacteroides generate some toxin and metabolite (phenol, indole) which later absorbed in blood and filter by kidney. Plasma metabolites of hemodialysis patients confirms the Colonic origin of indoxyl sulfate and p-cresol which is fermented product of phenylalanine and tyrosine.167
Factor that influence gut microbiota
Several intrinsic and extrinsic factors can influence gut microflora composition, some extrinsic factors like Diet/lifestyle, Drugs/Antibiotic, Age, Host immunity, and intrinsic factor involved genetic factors.168 There is some most studied factor that has been proven to alter gut microbiota.
Diet
In our routine life diet that we consume daily affect the microbe health in our gut. Reports showed that changing of fooding habits alter the microbe makeup in gut. Previously a mice study showed decline in gut microbial diversity after having high fat diet (60%). In diabetic patient A. muciniphila decreases, treatment with A. muciniphila reduces fat content. Among different kind of diets fiber rich diet is proven to be beneficial and associated with gut microbiota modulation.168 Composition of gut microbe varies to the consumed diet; Bacteroides composition is associated with protein and high animal fat diet. On the other hand Prevotella is more common to carbohydrate diet. Hence diet is associated with enterotype partitioning. The high-fat diet contributes to dysbiosis which ultimately caused disease.169 Strong health benefits evidence of fermented foods and beverages are reported in several diseases with their significance on the gut microbiota balance.170 Incorporation of fermented foods diet (e.g., kimchi, kefir, etc.) may counter the proinflammatory effects of gut dysbiosis.171
Infections
Some pathogenic microbe’s secreted metabolites can inhibit gut microbes growth. In a study where researcher showed Citrobacter rodentium enteropathogenic effect on the mice microbiota, reduced abundance of lactobacillus. Clostridium Difficile presence cause severe dysbiosis in gut microflora. In human infected patients microbiota studies in mice data showed transplantation results in high microbial richness and diversity. It is clearly proven by previous results that shift in the host microbiota have potential to clear infections and pathogenesis as well.
Medications
When people have any symptom and disease complication various medicines recommended in response to cure illness but some of them cure the disease but affecting the gut bacterial diversity. Antibiotics are the most commonly used class of drug prescribed for any type of infection and illness but showed a profound effect also, which could be persistent. Antibiotic consumption in neonate cause symbiosis, which may be an inclining factor to inflammatory bowel disease and obesity.172
Genetics
Host, microbiota and environment contribute or affect the genetic makeup of species. Presence of gut bacteria is influenced by genetic makeup affecting host metabolism and causing disease.173 As like common genetic makeup sibling or parent also showed similar microbiota communities than unrelated individuals. In monozygotic twins, gut microbiota is reported to be similar to dizygotic twins as genotyping and metagenome analysis showed.174
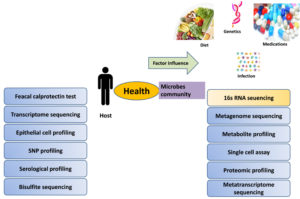
Diversity of microbes in gut allows degrading the complex constituents of diet and executing numerous vital activities that are essential for metabolic and immune functions. Intestine outer layer secrete mucin for protection from proinflammatory compounds and uptake of antigens, gut microbiota has long been believed to maintain, intestinal structure and cells function for providing better health.175 Butyrate is the metabolite produced by obligate anaerobic bacteria which stimulates mucin secretion from intestinal surface along with antimicrobial peptide; this secretion is as unique as your fingerprint. In 2007 NIH supported human microbiome project launched aiming, gut microbiome research experimental methodologies, new computational/statistical approaches for microbiome data analysis or a microbiome-based product or device. Past and present research implications confirm the role of microbiota to all aspect of development and host’s growth. Different practical efforts showed that imbalance in composition, habitat, or numbers of the gut microbiota tend to dysfunction in human is absolutely correlated. Many unsolved mysteries about microbe still remain unclear in the interactions study of host and the microbiota and their impact on the disease process. It is new field full of new promises, idea and unsolved biological questions that can expand knowledge in understanding of human pathophysiology. High-throughput sequencing data provides more information which support microbiology techniques and bioinformatics platforms in elucidating mechanisms of microbial community impact on human health. Study on human microbes can only be possible with current available machine based techniques and computer programmer. Microbes identity can be distinguishing with the help of bacterial ribosomal RNA (rRNA) genes sequencing, 16s ribosomal RNA is highly conserved gene among prokaryotes.176 Modern laboratory tools employed 16s rRNA and metagenomic sequencing enabled us to gain quantitative identification of microbes present in a sample (Figure 3). Phylogenetic information of microbiome attained using shotgun DNA sequencing approach, along with additional understanding of complex functions of these communities by identifying their genes. Customization of microbial consortia using advanced culturomics allows microbes interaction studies under complex system. Artificial intelligence in leading so many predictability in study field like microbe composition could predict age. Microbiome sequencing data obtained from fecal (gut microbiota), saliva (oral microbiota) and skin samples from several continents has been processed with bioinformatics pipeline by IBM researcher to ensure coherence and compatibility of the data.177 Hence gut microbes proved to be a health modulator, more researchers from different scientific background focus on how this advanced technology will continue decoding the gut microbiota potential in personalized medicine development.
Figure 3. Tools and techniques employed in host parameter and microbial diversity detection during disease pathogenesis along with influenced factors of gut microbial diversity
Characterization of oral cavity, skin microbiota may help us to better understand the complex gut microbiome-host interactions. International Human Microbiome Consortium (IHMC) provides a platform to access, data linking, and microbial sequences with human phenotype. These data bases and bioinformatics tools enables researcher to segregate diverse population and target potential groups for interventions. Human genome shape the structure of gut microbial composition. Expression of lactase gene responsible for lactose digestion has high Bifidobacteria, and Ruminococcus torques which correlates to fucosyl transferase (FUT2) gene.178 By microbe and genome interaction important metabolic pathways regulated in host that are important aspects of nutrition and immunity. So human disease and bacterial species analysis stretch can establish interrelation within microbes and host. Bifidobacterium abundance lowered the risk for inflammatory bowel disease ulcerative colitis, this observation also reported in previous clinical trials. Food majorly impact structure design of gut microbiome. Type of food and dietary patterns directly influences the prevalence of different types of bacteria in the gut, which affect human health through modulating metabolic pathways. It could be said that you feed your microbiota and are fed by them. Consumption of plant-based foods diet is more prone for developing “good” gut microbes. While processed plant food are associated with bad gut microbes. Mice feeds on the preservative component which are common to packed food (food emulsifier, processed sugar carboxymethylcellulose, polysorbate-80) showed enhanced expression of proinflammatory cytokine genes along with decrease Bacteroidales and Verrucomicrobia which promote mucus associated inflammation.179,180 Laxative drugs like progesterone, rupatadine and TNF-a inhibitors changed 10% of community variation in Dutch-Belgian population. Prebiotics are microbes growth promoting food such as fiber-rich foods, fruits, vegetables, and whole grains, while probiotic is bacterium or fungi that are taken as a supplement for proper body function, that occur in many fermented foods, including yogurt, sauerkraut, and tempeh. Consumption of fermented food gets you the live microbes that are present in them and improve intestinal permeability, balancing and barrier function. Dysbiosis is the cause of many disorders and reestablishment of symbiosis by introducing microbes could hold the key to new effective therapies. Probiotic or prebiotic interventions serve to increase beneficial microorganism’s growth and metabolism in the host. Restoration of the gut microbiota with the use of probiotics has led to promising results in clinical studies.. Microbiomics has incited interest pre/probiotics for management and control human health. Probiotics are reported to lower the inflammations which are reported to be elevated IgE, IgA, IL-10 and CRP. Probiotics, prebiotics, and their combinations clinically proved effective against gut disorders like IBD, digestion, traveller’s diarrhea.
Emerging areas of research provide promising cancer, brain, kidney, and obesity management since tools for probiotic research are now available. Therefore probiotics, prebiotics open up a new branch of science, flagging a new way toward personalized medicine, and future biotherapeutics. Phytochemicals consumption improve gut barrier integrity by activating tight junction expression and serving as antioxidant to induce stress resistance mechanisms in the gut by maintaining homeostasis, including autophagy, DNA repair, and expression of detoxifying and antioxidant enzymes., that helps to prevent chronic diseases, and prolongs lifespan. Phytochemicals that provide immunomodulatory potential first strengthen the gut system as gut is site of high immunological active cells. Much more emphasis should be given to phytochemical-microbiota reciprocal interactions, biotransformation of phytochemicals and plant-derived drugs, and pre-clinical and clinical efficacies of herbal medicine on dysbiosis. This will provide newer insights for future pre-clinical and clinical phytopharmacological interventions.
Based on experimental evidences companies are focusing on production of product help to establish healthy gut ecosystem. Probiotics supplemented food products cover $7.10 billion market by 2026 and increase of market based therapeutic in 2019 compared to 2018. Therapeutic marked goes down in 2020 due to covid-19 outbreak. Along with natural state microbes, other therapies also working on genetically-modified microbe. Microbiota being transferred via enema, colonoscopy, pills and nasogastric routes in patients. Keeping the benefits in mind National Microbiome Initiative set up goal to support microbial research and expanding it to pharma companies. Johnson & Johnson, Nestle, Seres Therapeutics, named little precision / personalized medicine which includes the microbiome, more companies are also working toward this approach.
Wellbeing role of microbiota open a window of nutritional and pharmacological opportunities to improve host-microbe relationship. How we respond toward certain drugs by enzymatic conversion it also depend on microbial type. In current world bacteriophage therapy is the only used in certain cases for patients in therapeutic failure, and are always accompanied by antibiotic treatment. Emerging phage therapy approach, which comprises phages and their derived protein against multi drug resistant bacteria. Phage lysins are more feasible therapeutic tool as they are not immunologically active and purification and production are accessible with safe storage. Extensive progress in last years has given deep insight to immunology and disease pathology with increased therapeutic pathways/targets exploration opportunity.
Gut microbes form the largest complex ecosystem in the human body which benefits the host by producing cometabolites. These metabolites depend on the type of food consumed by the host and also determine the microbiota composition, which later participates in vital activities connecting organs and mental health. Scientific interests focused on exploring more about microbes relation to host and their diseases got increased in the last years. Manifestation of fecal microbiota transfer, phage therapy, prebiotics, and probiotics approaches are getting success in controlling disease. Along with diet, other internal and external factors also disturb gut microflora composition that causes non-communicable diseases or increase susceptibility to communicable diseases. A short-chain fatty acid-containing diet regulates immune cell migration, mucosal permeability, and anti-inflammatory response and maintains a redox environment. More research carried out on the relationship between diet and host health provided evidence of an important role in human physiology and health in terms of pathogen invasion prevention and immune system maturation. Microbiota studies provide a base to design personalized medicine targeting gut microbiota for the ailment of a range of disorders in near future. Researchers have developed GI bacteria culturing techniques and metagenomic sequencing to overcome the limitation in this area. Microbiome ecology inspired use of live therapeutics to treat patients through additive, subtractive, and modulatory therapies. Pharma companies should focus on manufacturing bacteria-specific products having proper strain genomic characterization and minimal side effects. Functional network of these discipline come on front and provide a remarkable opportunities for diverse research avenues, emphasizing human medicine and biotechnology.
ACKNOWLEDGMENTS
None.
FUNDING
None.
DATA AVAILABILITY
Not applicable.
ETHICS STATEMENT
Not applicable.
- Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104-107.
Crossref - Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9(1):95.
Crossref - Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Moller K. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol. 2002;68(2):673-690.
Crossref - Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23(3):353-360.
Crossref - Min YW, Rhee P-L. The role of microbiota on the gut immunology. Clin Ther. 2015;37(5):968-975.
Crossref - de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017;152(1):1-12.
Crossref - Tamboli CP, Neut C, Desreumaux P, Colombel J. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1-4.
Crossref - Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microbial Ecol Health Dis. 2015;26(1):26191.
Crossref - Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004;9(2):180-197.
- Peterson J, Garges S, Giovanni M, et al. The NIH human microbiome project. Genome Research. 2009;19(12):2317-2323.
Crossref - Apajalahti J, Kettunen A. Microbes of the chicken gastrointestinal tract. Avian Gut Function in Health and Disease. 2006;28:107-123.
Crossref - Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65(10):4506-4512.
Crossref - Matsuki T, Watanabe K, Fujimoto J, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70(1):167-173.
Crossref - Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis. 2001;1(2):101-114.
Crossref - Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035s-145s.
Crossref - Ohland CL, MacNaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G807-G19.
Crossref - Tlaskalova-Hogenova H, Stepankova R, Hudcovic T, et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93(2-3):97-108.
Crossref - Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28(4):405-440.
Crossref - Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infect Immun. 2008;76(8):3360-3373.
Crossref - Zhang Y-J, Li S, Gan R-Y, Zhou T, Xu D-P, Li H-B. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16(4):7493-7519.
Crossref - Zakharchuk O, Kadelnik L, Kryvchanska M, Chokan V, Zakharchuk T. Gastrointestinal microflora and factors affecting intestinal normal flora in chronic dermatoses. Deutscher Wissenschaftsherold German Science Herald. 2019:3.
- Moll J, Wright V. Psoriatic arthritis. Seminars in arthritis and rheumatism; Elsevier. 1973:55-78.
Crossref - Sikora M, Stec A, Chrabaszcz M, et al. Gut Microbiome in Psoriasis: An Updated Review. Pathogens. 2020;9(6):463.
Crossref - Firestein GS, McInnes IB. Immunopathogenesis of rheumatoid arthritis. Immunity. 2017;46(2):183-196.
Crossref - Fox DA. The role of T cells in the immunopathogenesis of rheumatoid arthritis. Arthritis Rheum. 1997;40(4):598-609.
Crossref - Alesa DI, Alshamrani HM, Alzahrani YA, Alamssi DN, Alzahrani NS, Almohammadi ME. The role of gut microbiome in the pathogenesis of psoriasis and the therapeutic effects of probiotics. J Family Med Prim Care. 2019;8(11):3496-3503.
Crossref - Chen G, Chen Z-M, Fan X-Y, et al. Gut-Brain-Skin Axis in Psoriasis: A Review. Dermatol Ther. 2020;11(1):25-38.
Crossref - Myers B, Brownstone N, Reddy V, et al. The gut microbiome in psoriasis and psoriatic arthritis. Best Pract Res Clin Rheumatol. 2019;33(6):101494.
Crossref - Benhadou FMD, Mintoff D, Schnebert B, Thio HB. Psoriasis and Microbiota: A Systematic Review. Diseases. 20182;6(2):47.
Crossref - Groeger D, O’Mahony L, Murphy EF, et al. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut microbes. 2013;4(4):325-339.
Crossref - Holowacz S, Blondeau C, Guinobert I, Guilbot A, Hidalgo S, Bisson J-F. Lactobacillus salivarius LA307 and Lactobacillus rhamnosus LA305 attenuate skin inflammation in mice. Beneficial Microbes. 2018;9(2):299-309.
Crossref - Chen Y-H, Wu C-S, Chao Y-H, et al. Lactobacillus pentosus GMNL-77 inhibits skin lesions in imiquimod-induced psoriasis-like mice. J Food Drug Anal. 2017;25(3):559-566.
Crossref - Shepherd N. Pathological mimics of chronic inflammatory bowel disease. J Clin Pathol. 1991;44(9):726-733.
Crossref - Garrett WSPS, Gallini CA, Michaud M, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16(3):208-219.
Crossref - Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(2):327-39. e4.
Crossref - McDonald C, Inohara N, Nuñez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280(21):20177-80.
Crossref - Henckaerts L, Pierik M, Joossens M, Ferrante M, Rutgeerts P, Vermeire S. Mutations in pattern recognition receptor genes modulate seroreactivity to microbial antigens in patients with inflammatory bowel disease. Gut. 2007;56(11):1536-1542.
Crossref - Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79.
Crossref - Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(1):179-184.
Crossref - Chiaro TR, Soto R, Zac Stephens W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9(380):eaaf9044.
Crossref - Netea MGKB, de Jong DJ, Franke B, et al. NOD2 mediates anti-inflammatory signals induced by TLR2 ligands: implications for Crohn’s disease. Eur J Immunol. 2004;34(7):2052-2059.
Crossref - Couturier-Maillard AST, Rehman A, Normand S, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123(2):700-711.
Crossref - Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492-506.
Crossref - Packey CD, Sartor RB. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J Intern Med. 2008;263(6):597-606.
Crossref - Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, Stolte M. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 1997;11(5):853-858.
Crossref - Ramakrishna C, Kujawski M, Chu H, Li L, Mazmanian SK, Cantin EM. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat Commun. 2019;10(1):2153.
Crossref - Brun P, Castagliuolo I, Di Leo V, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G518-25.
Crossref - Iliev ID, Funari VA, Taylor KD, Nguyen Q, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314-1317.
Crossref - Etienne-Mesmin L, Chassaing B, Gewirtz AT. Tryptophan: a gut microbiota-derived metabolites regulating inflammation. World J Gastrointest Pharmacol Ther. 2017;8(1):7-9.
Crossref - Khan I, Ullah N, Zha L, et al. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens. 2019;8(3):126.
Crossref - Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis. 2010;16(4):684-95.
Crossref - Saemann MD, Bohmig GA, Osterreicher CH, Burtscher H, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14(15):2380-2382.
Crossref - Nagano Y, Itoh K, Honda K. The induction of Treg cells by gut-indigenous Clostridium. Curr Opin Immunol. 2012;24(4):392-397.
Crossref - Imaoka A, Shima T, Kato K, et al. Anti-inflammatory activity of probiotic Bifidobacterium: enhancement of IL-10 production in peripheral blood mononuclear cells from ulcerative colitis patients and inhibition of IL-8 secretion in HT-29 cells. World J Gastroenterol. 2008;14(16):2511-2516.
Crossref - Benjamin JL, Hedin CRH, Koutsoumpas A, et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18(6):1092-1100.
Crossref - Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489-1499.
Crossref - Compare D, Coccoli P, Rocco A, et al. Gut-liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22(6):471-476.
Crossref - Malaguarnera G, Giordano M, Nunnari G, Bertino G, Malaguarnera M. Gut microbiota in alcoholic liver disease: pathogenetic role and therapeutic perspectives. World J Gastroenterol. 2014;20(44):16639-16648.
Crossref - Machado MV, Cortez-Pinto H. Gut microbiota and nonalcoholic fatty liver disease. Ann Hepatol. 2012;11(4):440-449.
Crossref - de Faria Ghetti F, Oliveira DG, de Oliveira JM, De Castro LEVV, Cesar DE, Moreira APB. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr. 2018;57(3):861-876.
Crossref - Yu L-X, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14(9):527-539.
Crossref - Gerussi A, Luca M, Cristoferi L, et al. New therapeutic targets in autoimmune cholangiopathies. Front Med. 2020;7:117.
Crossref - Thuy S, Ladurner R, Volynets V, et al. Nonalcoholic fatty liver disease in humans is associated with increased plasma endotoxin and plasminogen activator inhibitor 1 concentrations and with fructose intake. J Nutr. 2008;138(8):1452-1455.
Crossref - Jarrar M, Baranova A, Collantes R, et al. Adipokines and cytokines in non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2008;27(5):412-421.
Crossref - Tsuei J, Chau T, Mills D, Wan Y-JY. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med. 2014;239(11):1489-1504.
Crossref - Suresh MV, Singh SK, Ferguson DA, Agrawal A. Human C-reactive protein protects mice from Streptococcus pneumoniae infection without binding to pneumococcal C-polysaccharide. J Immunol. 2007;178(2):1158-1163.
Crossref - Thomas-Rudolph D, Du Clos TW, Snapper CM, Mold C. C-reactive protein enhances immunity to Streptococcus pneumoniae by targeting uptake to FcγR on dendritic cells. J Immunol. 2007;178(11):7283-7291.
Crossref - Mortensen RF. C-reactive protein, inflammation, and innate immunity. Immunologic Research. 2001;24(2):163-176.
Crossref - Shasthry SM. Fecal microbiota transplantation in alcohol related liver diseases. Clin Mol Hepatol. 2020;26(3):294-301.
Crossref - Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601-609.
Crossref - Chen Y, Yang F, Lu H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562-572.
Crossref - Esposito E, Iacono A, Bianco G, et al. Probiotics reduce the inflammatory response induced by a high-fat diet in the liver of young rats. J Nutr. 2009;139(5):905-911.
Crossref - Aballay LR, Eynard AR, Diaz MdP, Navarro A, Munoz SE. Overweight and obesity: a review of their relationship to metabolic syndrome, cardiovascular disease, and cancer in South America. Nutr Rev. 2013;71(3):168-179.
Crossref - Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262-266.
Crossref - Goran MI, Ball GD, Cruz ML. Obesity and risk of type 2 diabetes and cardiovascular disease in children and adolescents. J Clin Endocrinol Metab. 2003;88(4):1417-1427.
Crossref - Calder PC. Omega-3 polyunsaturated fatty acids and inflammatory processes: nutrition or pharmacology? Br J Clin Pharmacol. 2013;75(3):645-662.
Crossref - Lackey DE, Lynch CJ, Olson KC, et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am J Physiol Endocrinol Metab. 2013;304(11):E1175-E1187.
Crossref - Longo M, Zatterale F, Naderi J, et al. Adipose tissue dysfunction as determinant of obesity-associated metabolic complications. Int J Mol Sci. 2019;20(9):2358.
Crossref - Chen J, Li Y, Tian Y, et al. Interaction between Microbes and Host Intestinal Health: Modulation by Dietary Nutrients and Gut-Brain-Endocrine-Immune Axis. Curr Protein Pept Sci. 2015;16 (7):592-603.
Crossref - Munoz-Garach A, Diaz-Perdigones C, Francisco J. Tinahones. Gut microbiota and type 2 diabetes mellitus. Endocrinologia y Nutricion. 2016;63(10):560-568.
Crossref - Vrieze A, Nood EV, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143 (4):913-6.e7.
Crossref - Pedersen H, Guomundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376-381.
Crossref - Ottosson F, Brunkwall L, Ericson U, et al. Connection Between BMI-Related Plasma Metabolite Profile and Gut Microbiota. J Clin Endocrinol Metab. 2018;103(4):1491-1501.
Crossref - Zhou K. Strategies to promote abundance of Akkermansia muciniphila aepitg, evidence from dietary intervention studies. J Funct Foods. 2017;33:194-201.
Crossref - Roshanravan N, Bastani S, Tutunchi H, et al. A comprehensive systematic review of the effectiveness of Akkermansia muciniphila, a member of the gut microbiome, for the management of obesity and associated metabolic disorders. Arch Physiol Biochem. 2021:1-11.
Crossref - Schroeder BO, Birchenough GMH, Stahlman M, et al. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe. 2018;23(1):27-40.e7.
Crossref - Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124(10):4204-4211.
Crossref - Leustean AM, Ciocoiu M, Sava A, et al. Implications of the intestinal microbiota in diagnosing the progression of diabetes and the presence of cardiovascular complications. J Diabetes Res. 2018;5205126.
Crossref - Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4(1):15.
Crossref - Kirichenko TV, Markina YV, Sukhorukov VN, Khotina VA, Wu W-K, Orekhov AN. A novel insight at atherogenesis: the role of microbiome. Front Cell Dev Biol. 2020;8:586189.
Crossref - Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576-585.
Crossref - Geng J, Yang C, Wang B, et al. Trimethylamine N-oxide promotes atherosclerosis via CD36-dependent MAPK/JNK pathway. Biomed Pharmacother. 2018;97:941-947.
Crossref - Brocheriou I, Maouche S, Durand H, et al. Antagonistic regulation of macrophage phenotype by M-CSF and GM-CSF: implication in atherosclerosis. Atherosclerosis. 2011;214(2):316-324.
Crossref - Pieczynska MD, Yang Y, Petrykowski S, Horbanczuk OK, Atanasov AG, Horbanczuk JO. Gut Microbiota and Its Metabolites in Atherosclerosis Development. Molecules. 2020;25(3):594.
Crossref - Elfaki I, Mir R, Almutairi FM, Duhier FMA. Cytochrome P450: polymorphisms and roles in cancer, diabetes and atherosclerosis. Asian Pac J Cancer Prev. 2018;19(8):2057-2070.
- Sima AV, Stancu CS, Simionescu M. Vascular endothelium in atherosclerosis. Cell Tissue Res. 2009;335(1):191-203.
Crossref - Rayner KJ, Sheedy FJ, Esau CC, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921-2931.
Crossref - Ohashi R, Mu H, Wang X, Yao Q, Chen C. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. Qjm. 2005;98(12):845-856.
Crossref - Chen PB, Black AS, Sobel AL, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol. 2020;38(11):1288-1297.
Crossref - Liu S, Zhao W, Liu X, Cheng L. Metagenomic analysis of the gut microbiome in atherosclerosis patients identify cross-cohort microbial signatures and potential therapeutic target. The FASEB Journal. 2020;34(11):14166-14181.
Crossref - Jie Z, Xia H, Zhong S-L, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845.
Crossref - Miremadi F. Hypocholesterolemic and Anti-hypertensive Properties of Lactobacilli and Bifidobacteria: Victoria University; 2018.
- Li F, Zhang T, He Y, et al. Inflammation inhibition and gut microbiota regulation by TSG to combat atherosclerosis in ApoE−/− mice. J Ethnopharmacol. 2020;247:112232.
Crossref - Chittim CL, Irwin SM, Balskus EP. Deciphering Human Gut Microbiota-Nutrient Interactions: A Role for Biochemistry. Biochemistry. 2018;57(18):2567-2577.
Crossref - Duda-Chodak A TT, Satora P, Sroka P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur J Nutr. 2015;54(3):325-341.
Crossref - Khurana S VK, Hollingsworth A, Piche M, Tai TC. Polyphenols: benefits to the cardiovascular system in health and in aging. Nutrients. 2013;5(10):3779-827.
Crossref - Sharma R, Padwad Y. Probiotic bacteria as modulators of cellular senescence: emerging concepts and opportunities. Gut Microbes. 2020;11(3):335-349.
Crossref - Toward RE, Walton GE, Gibson GR. Immunosenescence and the gut microbiota: the role of probiotics and prebiotics. Nutrition and Aging. 2012;1(3-4):167-180.
Crossref - Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9(2):107-116.
Crossref - Macia L, Thorburn AN, Binge LC, et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev. 2012;245(1):164-176.
Crossref - Buford TW. (Dis) Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5(1):80.
Crossref - Franceschi C, Capri M, Monti D, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128(1):92-105.
Crossref - Kim K-A, Jeong J-J, Yoo S-Y, Kim D-H. Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC Microbiol. 2016;16(1):9.
Crossref - Stehle Jr JR, Leng X, Kitzman DW, Nicklas BJ, Kritchevsky SB, High KP. Lipopolysaccharide-binding protein, a surrogate marker of microbial translocation, is associated with physical function in healthy older adults. J Gerontol A Biol Sci Med Sci. 2012;67(11):1212-1218.
Crossref - Hersoug LG, Moller P, Loft S. Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: implications for inflammation and obesity. Obes Rev. 2016;17(4):297-312.
Crossref - Mukherjee D, Chora AF, Mota MM. Microbiota, a third player in the host-Plasmodium affair. Trends Parasitol. 2020;36(1):11-18.
Crossref - Nash AA, Dalziel RG, Fitzgerald JR. Mims’ pathogenesis of infectious disease: Academic Press; 2015.
- Chen C-C, Louie S, McCormick B, Walker WA, Shi HN. Concurrent infection with an intestinal helminth parasite impairs host resistance to enteric Citrobacter rodentium and enhances Citrobacter-induced colitis in mice. Infect Immun. 2005;73(9):5468-5481.
Crossref - Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119-129.
Crossref - Yilmaz B, Portugal S, Tran TM, et al. Gut microbiota elicits a protective immune response against malaria transmission. Cell. 2014;159(6):1277-1289.
Crossref - Flippo TS, Holder WD. Neurologic degeneration associated with nitrous oxide anesthesia in patients with vitamin B12 deficiency. Arch Surg. 1993;128(12):1391-1395.
Crossref - Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327-336.
Crossref - Issac TG SS, Christopher R, Chandra SR. Vitamin B12 deficiency: an important reversible co-morbidity in neuropsychiatric manifestations. Indian J Psychol Med. 2015;37(1):26-29.
Crossref - Roth JR LJ, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137-181.
Crossref - Xu Y, Xiang S, Ye K, et al. Cobalamin (Vitamin B12) Induced a Shift in Microbial Composition and Metabolic Activity in an in vitro Colon Simulation. Front Microbiol. 2018;9:2780.
Crossref - Khan MT BW, van Dijl JM, Harmsen HJ. How can Faecalibacterium prausnitzii employ riboflavin for extracellular electron transfer? Antioxid Redox Signal. 2012;17(10):1433-1440.
Crossref - Iannone LFPA, Blottiere HM, Clarke G, et al. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev Neurother. 2019;19(10):1037-1050.
Crossref - Morris G, Berk M, Carvalho AF, Caso JR, Sanz Y, Maes M. The Role of Microbiota and Intestinal Permeability in the Pathophysiology of Autoimmune and Neuroimmune Processes with an Emphasis on Inflammatory Bowel Disease Type 1 Diabetes and Chronic Fatigue Syndrome. Curr Pharm Des. 2016;22(40):6058-6075.
Crossref - Ma Q XC, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. 2019;16(1):53.
Crossref - Stilling RM, vdWouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochem Int. 2016;99:110-132.
Crossref - Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91-119.
Crossref - Vinolo MA RH, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3(10):858-876.
Crossref - Liu X-F, Zhou D-D, Hao J-L, et al. The Nrf2 signaling in retinal ganglion cells under oxidative stress in ocular neurodegenerative diseases. Int J Biol Sci. 2018;14(9):1090-1098.
Crossref - Lv CMS, Wang Q, Sun Y, et al. a-Lipoic Acid Promotes Neurological Recovery After Ischemic Stroke by Activating the Nrf2/HO-1 Pathway to Attenuate Oxidative Damage. Cell Physiol Biochem. 2017;43(3):1273-1287.
Crossref - Natale G BF, Busceti CL, Gambardella S, Limanaqi F, Fornai F. TREM Receptors Connecting Bowel Inflammation to Neurodegenerative Disorders. Cells. 2019;8(10):1124.
Crossref - Raval U, Harary JM, Zeng E, Pasinetti GM. The dichotomous role of the gut microbiome in exacerbating and ameliorating neurodegenerative disorders. Expert Rev Neurother. 2020;20(7):673-686.
Crossref - Zhu S, Jiang Y, Xu K, et al. The progress of gut microbiome research related to brain disorders. J Neuroinflammation. 2020;17(1):25.
Crossref - McLean G HJ, Guthrie B, Mercer SW. Co-morbidity and polypharmacy in Parkinson’s disease: insights from a large Scottish primary care database. BMC Neurol. 2017;17(1):126.
Crossref - Limbana T KF, Eskander N. Gut Microbiome and Depression: How Microbes Affect the Way We Think. Cureus. 2020;12(8):e9966.
Crossref - Zigmond M, Clayton W, Chesselet M-F, eds. Neurobiology of brain disorders: biological basis of neurological and psychiatric disorders. Academic press, 2022.
- Jiang C LG, Huang P, Liu Z, Zhao B. The Gut Microbiota and Alzheimer’s Disease. J Alzheimers Dis. 2017;58(1):1-15.
Crossref - Wasser CI, Mercieca E, Kong G, et al. Gut dysbiosis in Huntington’s disease: associations among gut microbiota, cognitive performance and clinical outcomes. Brain Commun. 2020;2(2):fcaa110.
Crossref - Tikale, Swati, et al. Management of lumber canal stenosis through Panchakarma: Case study. Indian Journal of Forensic Medicine & Toxicology. 2020;14(4): 6381-6385.
Crossref - Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci. 2015;9:392.
Crossref - Szczesniak O HK, Hanssen JF, Rudi K. Isovaleric acid in stool correlates with human depression. Nutr Neurosci. 2016;19(7):279-83.
Crossref - Kelly JR, Allen AP, Temko A, et al. Lost in translation? The potential psychobiotic Lactobacillus rhamnosus (JB-1) fails to modulate stress or cognitive performance in healthy male subjects. Brain Behav Immun. 2017;61:50-59.
Crossref - Karakula-Juchnowicz H, Rog J, Juchnowicz D, et al. The study evaluating the effect of probiotic supplementation on the mental status, inflammation, and intestinal barrier in major depressive disorder patients using gluten-free or gluten-containing diet (SANGUT study): a 12-week, randomized, double-blind, and placebo-controlled clinical study protocol. Nutr J. 2019;18(1):50.
Crossref - Huang R, Wang K, Hu J. Effect of Probiotics on Depression: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients. 2016;8(8):483.
Crossref - Organization WH. Global action plan on physical activity 2018-2030: more active people for a healthier world: World Health Organization; 2019.
- Vivarelli S, Salemi R, Candido S, et al. Gut microbiota and cancer: from pathogenesis to therapy. Cancers. 2019;11(1):38.
Crossref - Gamallat Y, Meyiah A, Kuugbee ED, et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed Pharmacother. 2016;83:536-541.
Crossref - Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743.
Crossref - Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumor Biol. 2013;34(3):1285-300.
Crossref - Wang L, Yu K, Zhang X, Yu S. Dual functional roles of the MyD88 signaling in colorectal cancer development. Biomed Pharmacother. 2018;107:177-184.
Crossref - Gianotti L, Morelli L, Galbiati F, et al. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol. 2010;16(2):167.
Crossref - Cianci R, Franza L, Schinzari G, et al. The interplay between immunity and microbiota at intestinal immunological niche: the case of cancer. Int J Mol Sci. 2019;20(3):501.
Crossref - Pere-Vedrenne C, Cardinaud B, Varon C, et al. The cytolethal distending toxin subunit CdtB of Helicobacter induces a Th17-related and antimicrobial signature in intestinal and hepatic cells in vitro. J Infect Dis. 2016;213(12):1979-1989.
Crossref - Lin W-W, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117(5):1175-1183.
Crossref - von Hertzen LC, Joensuu H, Haahtela T. Microbial deprivation, inflammation and cancer. Cancer Metastasis Rev. 2011;30(2):211-223.
Crossref - Vaezi MF, Falk GW, Peek RM, et al. CagA-positive strains of Helicobacter pylori may protect against Barrett’s esophagus. Am J Gastroenterol. 2000;95(9):2206-2211.
Crossref - Fischbach LA, Graham DY, Kramer JR, et al. Association between Helicobacter pylori and Barrett’s Esophagus: A Case-Control Study. Am J Gastroenterol. 2014;109(3):357.
Crossref - Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83(2):308-315.
Crossref - Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25(4):657-670.
Crossref - Jovanovich A, Isakova T, Stubbs J. Microbiome and Cardiovascular Disease in CKD. Clin J Am Soc Nephrol. 2018;13(10):1598-1604.
Crossref - Simeoni M, Citraro ML, Cerantonio A, et al. An open-label, randomized, placebo-controlled study on the effectiveness of a novel probiotics administration protocol (ProbiotiCKD) in patients with mild renal insufficiency (stage 3a of CKD). Eur J Nutr. 2019;58(5):2145-2156.
Crossref - Castillo-Rodriguez E, Fernandesz-Prado R, Esteras R, et al. Impact of Altered Intestinal Microbiota on Chronic Kidney Disease Progression. Toxins. 2018;10(7):300.
Crossref - Mafra D, Borges N, Alvarenga L, et al. Dietary Components That May Influence the Disturbed Gut Microbiota in Chronic Kidney Disease. Nutrients. 2019;11(3):496.
Crossref - Wen L, Duffy A. Factors Influencing the Gut Microbiota, Inflammation, and Type 2 Diabetes. J Nutr. 2017;147(7):1468S-1475S.
Crossref - He C, Cheng D, Peng C, Li Y, Zhu Y, Lu N. High-Fat Diet Induces Dysbiosis of Gastric Microbiota Prior to Gut Microbiota in Association With Metabolic Disorders in Mice. Front Microbiol. 2018;9:639.
Crossref - Bell V, Ferrao J, Pimentel L, Pintado M, Fernandes T. One Health, Fermented Foods, and Gut Microbiota. Foods. 2018;7(12):195.
Crossref - Stiemsma LT, Nakamura RE, Nguyen JG, Michels KB. Does Consumption of Fermented Foods Modify the Human Gut Microbiota? J Nutr. 2020;150(7):1680-1692.
Crossref - Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140(6):1729-1737.
Crossref - Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933-18938.
Crossref - Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417-421.
Crossref - Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313-323.
Crossref - Rajendhran J, Gunasekaran P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res. 2011;166(2):99-110.
Crossref - Huang CF, Zhang L, Ma SR, et al. Clinical significance of Keap1 and Nrf2 in oral squamous cell carcinoma. PLoS One. 2013;8(12):e83479.
Crossref - Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156-165.
Crossref - Partridge D, Lloyd KA, Rhodes JM, Walker AW, Johnstone AM, Campbell BJ. Food additives: Assessing the impact of exposure to permitted emulsifiers on bowel and metabolic health-introducing the FADiets study. Nutr Bull. 2019;44(4):329-349.
Crossref - Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. Bmj. 2018;361.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.