ISSN: 0973-7510
E-ISSN: 2581-690X
Fungal infections, such as those caused by Aspergillus, Alternaria, Colletotrichum, Penicillium, and Rhizopus, are considered the major global threat to human life. In seeking a treatment, we synthesized and characterized copper nanoparticles (CuNPs) using Curcuma longa extract. C. longa plant extract has been previously studied and validated for its strong antimicrobial properties. Novel green particles were synthesized in this study using C. longa and copper nitrate. We also investigated antifungal activity through inhibition studies and real-time expression of gene members belonging to the chitin synthase family. Synthesized CuNPs were characterized by ultraviolet-visible spectroscopy, Fourier transform-infrared spectroscopy, X-ray diffraction, and nanoparticle tracking analysis. Gene expression was determined by real-time PCR. The results visibly confirmed the antifungal activity of the synthesized CuNPs against Aspergillus through zones of growth inhibition. The zone diameters were comparable to those of the positive control used in the study. The synthesized CuNPs were 60 nm in diameter and with a resonance peak at 535 nm. The observations of the downregulation of chitin synthase gene members 1, 2, and 3 suggest significant antifungal activity of the synthesized CuNPs. The collective findings indicate the potential value of green-synthesized CuNPs as antifungal agents.
Curcuma Longa, Antifungal, CuNPs, NTA, Real-time PCR, Chitin Synthase
Nanoscience is a relatively new field of science. In nanoscience, technology, engineering, and mathematical disciplines are involved in the creation of nanomaterials ranging in size from one to 100 nm.1 The manufacture of nanoparticles (NPs), which may be a better substitute for antibiotics, has been made possible by advances in nanotechnology. This has expanded the field of nanomedicine and given rise to new avenues for research.2 Numerous nanomaterials are being used in catalysis, medical equipment, gadgets, water, and food processing.3
Copper NPs (CuNPs) have drawn the most attention among the various metal-based NPs because they can be produced in greater abundance and have more useful features than the more popular silver or gold NPs. In addition to being used as fungicides, insecticides, algaecides, and herbicides in agriculture, copper compounds are widely used as disinfectants.4
The special properties of CuNPs, such as high surface-to-volume ratio and crystalline surface structure, results in CuNPs higher antibacterial activity than copper salts.5 CuNPs have potent inhibitory and antifungal effects.6,7
Turmeric (Curcuma longa) is a flowering plant that has been traditionally and widely used in food products. The antimicrobial effects of curcumin and turmeric on fungi-related spoilage and microbial infections have been demonstrated. Medicinal attributes of C. longa include insecticidal, antibiotic,8 antifungal,9 antimalarial, antiviral,10 and antioxidant activities.11
C. longa inhibits the growth of mycelia in fungi, which are responsible for the degradation of agricultural products.12 Both ethanol and hexane extracts of C. longa have significant antifungal activities against many pathogenic fungal strains, including Botrytis cinerea, Fusarium graminearum, and Sclerotinia sclerotiorum).13,14
Chitin synthases (CHSs) are important enzymes that catalyze N-acetylglucosamine polymerization.15,16 The enzymes, which typically reside in the cytoplasmic membrane of fungi, have generated much interest as potential targets of novel fungicides.17 Seven CHS genes have been identified in various fungi.18 The transmembrane and chitin synthase domains are shared by all CHSs. Chitin concentration and CHS composition vary among fungi.19 Three CHS genes (CHS1, 2, and 3) in S. cerevisiae each play a specific role in cell wall expansion and budding.20 CHS1 strengthens damaged cell walls of daughter cells following division. In primary septa, CHS2 catalyzes the production of chitin, which is necessary for both cell division and septum development.21 Chitin ring development at the base of developing buds and chitin production in the lateral cell depend on CHS3.22
In the current study, we aimed to synthesize CuNPs using C. longa extracts and screen for their potential antifungal activity directed at CHSs using preliminary inhibition assays and real-time quantification. We also characterized the synthesized CuNPs using ultraviolet (UV) spectroscopy, Fourier transform-infrared spectroscopy (FTIR), and NP tracking analysis (NTA).
Preparation of C. longa extracts: Tubers of C. longa were collected from a local greenhouse on the University of Agricultural Sciences Bangalore, GKVK campus. The tubers were cleaned to remove adhering mud particles and other impurities, sun-dried for more than a week, and finely pulverized. The dried powder (10 g) was extracted with methanol and ethanol. A total of 100 mL of solvent was used for extraction using the Soxhlet apparatus at 45°C. The extract was dried and stored until required. This extract was used as either a reducing or capping agent to synthesize the CuNPs.
Green synthesis: Copper nitrate solution (0.1M/100 mL) was added to 50 mL of previously prepared 10% C. longa extract. The contents were then magnetically stirred for approximately 2-3 days at room temperature. The color change of the solution confirmed the formation of CuNPs. The synthesized CuNPs were characterized concerning their morphology, particle size, and stability. These were used for antifungal activity and gene expression studies using real-time PCR.23
Test for antifungal activity: To estimate the antifungal activity of the prepared green NPs, microdilution was performed. Fungal spores were collected by thoroughly washing the surface of agar plates with sterile 0.85% saline and 0.1% (v/v) Tween 80. The fungal suspension was further standardized to 106 conidia/mL with sterile saline (0.85%) and approximately 100 μL of fungal suspension was spread on potato dextrose agar plates.24
Five millimeter diameter holes were created using a gel punch. Each hole was loaded with a samples. Dimethyl sulfoxide (DMSO) was used as the negative control. The fungicide bavistin (0.1%) was used as a positive control. The plates were incubated at (28 ± 2)°C for 3-4 days. Antifungal activity was screened by recording the diameter of the inhibition zone on the plate after 3-4 days.
Characterization of nanoparticles
The average size of the synthesized particles was measured by dynamic light scattering (DLS) and the polydispersity index (PDI) was determined using a model SZ-100 DLS device (Horiba) using scattered light at 90/173°angles (default). Zeta potential was measured at approximately 25°C to screen the dispersion and overall stability of the synthesized CuNPs. The absorbance was measured using a model 1800 UV spectrophotometer (Shimadzu, Japan) at wavelengths ranging from 190 to 800 nm. Control-cupric nitrate (CuNO3) salt along NP solutions were used.
The synthesized green CuNPs were analyzed at 500–4000 cm−1 using an FTIR spectrometer (Shimadzu) to identify biomolecules within the plant extracts. The size and distribution of the synthesized NPs were screened for their particle size and distribution by NTA using a model LM-20 apparatus (NanoSight Ltd., UK). NTA separates particles based on their size and intensity, and simultaneously determines their Brownian motion.
Treatment of Aspergillus with synthesized NPs
Potato dextrose broth (PDB) was used. Approximately 1×1 mm pieces of mycelium from the culture on agar plates were used to inoculate 50 mL of PDB) in 100 mL Erlenmeyer flasks. These cultures were allowed to incubated statically 4-5 days in an incubator (REMI, India) at 25±2°C before extraction as previously described.23 Flasks without NP treatment served as negative controls. Bavistin (1%) was used as a positive control. CuNO3 solution served as the negative control. Green CuNPs (10%) were used as the treatment. Following treatment, the mycelial mats were collected by filtration and used for gene expression studies.
RNA extraction
Total RNAs were extracted from 2-day-old mycelia.25 The harvested mycelia were ground to a fine powder with liquid N2. Two milliliters TRIzol Reagent (HiMedia, India) was added to the slurry. Following incubation at room temperature for 5 min, 300 μL of chloroform was added to the sample, centrifuged at 10.000 rpm for 15s, and further incubated at room temperature for 3 min. The contents were then centrifuged at 12,000 rpm for 15 min at 4°C. The RNA was precipitated from the obtained supernatant using 500 μL isopropanol at −20°C for 2h. The RNA was centrifuged at 13,000 rpm for 15 min at 4°C, washed with ice-cold 75% ethanol, and re-suspended in 20 μL of DNase-RNase free water. Synthesis of complementary DNA (cDNA) was performed using Superscript TMII Reverse Transcriptase (200 U/μL) with approximately 1.31 μL of RNA (obtained concentration of 1.75 μg/μL). Random primers along with 1 μL of reverse transcriptase were added and incubated at 25°C for 10 min. The cDNA obtained was used for real-time PCR analysis.
Real-time PCR
All the primers that were used were designed with Primer3 software and purchased from Eurofins Scientific (France). Real-time PCR was done as previously described26 using SYBR Green Supermix (HiMedia). Primers (600 nM each) were used, and the total volume maintained at 12.5 μL. Three independent experiments were done.
Expression of CHS gene members
qPCR was performed with samples (both control and treatment) in a thermocycler (Bio-Rad, USA) for approximately 40 cycles at 94°C for 50 s, 65°C for 50 s, and 72°C for 60 s. The housekeeping gene actin was also amplified along with the gene members to comparatively analyze mRNA levels (Table). Expression analysis was performed using the ΔΔCt method (Ct values for each member were normalized according to Actin D).
Table :
List of the primers that were used for the real time PCR.
| Gene | Template strand | Length | Tm | GC% | Product length | |
|---|---|---|---|---|---|---|
| CHS1 | FW | ATGAACAGCTGGCTAACCfor CC | 20 | 60.03 | 55 | 655 |
| RV | TAGATGTTCGTACCGCTCGC | 20 | 59.97 | 55 | ||
| CHS2 | FW | CCGCGATAAAAAGCTCCGTG | 20 | 59.97 | 55 | 842 |
| RV | CAATTGCGCATCCCGCATAA | 20 | 59.97 | 50 | ||
| CHS3 | FW | CCCGCTGCTATTGCATTCAC | 20 | 59.97 | 55 | 413 |
| RV | GCATCAGTCAGTACAAGCCCT | 21 | 60.07 | 52.38 | ||
| Actin | FW | GACCGACTACCTGATGAAGA | 20 | 58.65 | 50 | 450 |
| RV | TGCCGATGGTGATAACCTG | 19 | 58.97 | 50 |
FW: forward primer; RV: reverse primer. Tm: melting temperature
Statistical analysis
All values are expressed as the average (±standard deviation). The results were analyzed by one-way analysis of variance (ANOVA, p<0.05) using SPSS software (faculty version). Statistical significance was set at p < 0.05.
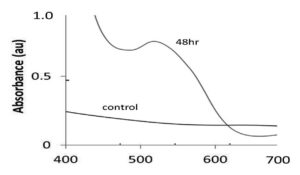
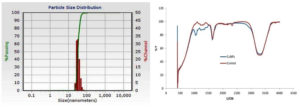
Visual observation was used as a preliminary screening method for the synthesis of CuNPs. Color change from dark brown to black color (Figure 1), was indicative of CuNP formation. CuNPs characterized by UV–vis spectrophotometry. The UV–vis spectra of 0.001M CuNO3 and the CuNP solution were observed immediately after synthesis at 48 h. Both exhibited a characteristic resonance peak at 535 nm (Figure 2). NTA images showed that the NPs had a size of 60 nm, which was calculated based on their Brownian motion. The synthesized CuNPs were subjected to FTIR analysis; two peaks were evident at 1720 and 3300cm-1 (Figure 3). NTA revealed the size distribution of the CuNPs with a mean diameter of 60 nm for the green CuNPs after 48 h of incubation (Figure 3). The zeta potential of the synthesized nanoparticles was -15.3, indicating that the synthesized nanoparticles do exhibited a high degree of stability. The total concentration of CuNPs measured by NTA at 24 h was almost 6.2-fold lower than that of samples after 48 h of incubation.
Figure 1. Representative images of copper nanoparticle dispersions synthesized with Curcuma longa extracts after 24hr and 48hr of reaction time. Control beakers contained copper nitrate solution without any treatment
Figure 2. Graph showing the UV–visible spectra of synthesized CuNPs after 48hr. Control is the Copper nitrate solution (0.01M)
Figure 3. Left: Image showing the size distribution of synthesized copper nanoparticles. Right: FTIR spectra of synthesized copper nanoparticles using Curcuma longa extracts after 48hr of incubation
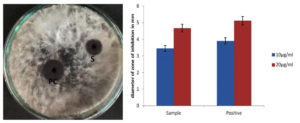
The synthesized CuNPs showed inhibition zone of 3.45±0.34 and 4.67±1.14 when applied at 10 and 20 µg/mL, respectively. The positive control showed zones of inhibition of 3.9±0.51 and 5.12±1.31 at 10 and 20 µg/mL, respectively (Figure 4). The antifungal activity of the samples was similar to that of the positive control. Keservani27 reported similar findings of high antifungal activity attributed to C. longa. The authors obtained promising antifungal activity with C. longa alcohol extract against many Fusarium and Alternaria species. Chen et al. also reported similar findings, where C. longa inhibited Aspergillus flavus and disrupted plasma membrane integrity.28
Figure 4. Plate showing the antifungal activity of CuNPs against Aspergillus culture on PDA media. PC: Positive control; S: Sample after 48hr of incubation. Right: Graph is showing the mean values of inhibition zone diameters that were observed at varying concentrations. All the experiments are done in triplicates
Differential expression of CHs gene members
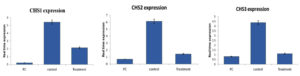
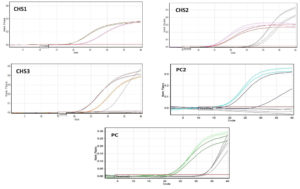
The differential expression of CHS genes were studied using qRT-PCR using RNA isolated from hyphae as the template. The expression levels of CHS1, CHS2, and CHS3 were compared in control and treated (CuNP) samples. The expression levels of CHS1, CHS2, and CHS3 were 4%, 11%, and 19%, respectively, for the positive control (bavistin) compared to the control (100%, without treatment) (Figure 5). The control was 100% for all three genes (p<0.05). CHS1 was expressed in 39% of NPs (p<0.05). The CHS2 gene was 23% overexpressed with treatment (p<0.05) and CHS3 was 26% overexpressed with treatment (p<0.05). Two-way ANOVA) was done to comparatively analyze the expression levels at p<0.05. A significant difference was observed in terms of the expression of all three genes between the treatment and control groups [F(2,4)=18.72505; p=0.0017]. However, no significant effect was observed between members [F(2,4)=0.3390; p=0.49739].
Figure 5. Real-time expression values before and after treatment with CuNPs. Relative expression of members was studied individually. The data was plotted after normalization with the expression of housekeeping gene actin. All the values are independent experiments and expressed as relative expression ± SD
According to Kong,29 CHS1, CHS6, and CHS7 aid in plant infection. The CHS1 mutant displayed an altered conidial morphology, making germination difficult. Fungal growth is reduced, along with virulence. The authors also observed a reduction in growth rate upon deletion of the CHS2 and CHS6 genes.29 Fluconazole-resistant colonies expressed high amounts of CHS genes compared to susceptible isolates. CHS3 expression was elevated ten times in strains resistant to fluconazole.30 The results of our study are almost identical to these findings. We observed that CHS1, 2, and 3 genes were significantly reduced expression with associated functional alterations.
We describe a simple and economical green synthesis method for CuNPs using C. longa plant extracts as reducing agents. The synthesized CuNPs/plant extracts could be potential antifungal agents against Aspergillus. We studied the optical and structural stability, and antifungal activity of CuNPs. DLS indicated sizes ranging from 50 to 60 nm. FTIR and NTA confirmed the presence of compounds around the CuNPs. CuNPs synthesized using C. longa extract exhibited promising antifungal activity against Aspergillus as validated by inhibition and quantitative PCR studies (Figure 6).
ACKNOWLEDGMENTS
None.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Aljamali NM, Thamer AK, Sabea AM. Review on electronic instruments and its nano-skill solicitations. Journal of Electrical and Power System Engineering. 2021;7(3):11-19.
- Mubeen B, Ansar AN, Rasool R, et al. Nanotechnology as a Novel Approach in Combating Microbes Providing an Alternative to Antibiotics. Antibiotics. 2021;10(12):1473.
Crossref - Aragaw TA, Bogale FM, Aragaw BA. Iron-based nanoparticles in wastewater treatment: A review on synthesis methods, applications, and removal mechanisms. Journal of Saudi Chemical Society. 2021;25(8):101280.
Crossref - Huang Q, Zhu Y. Printing conductive nanomaterials for flexible and stretchable electronics: A review of materials, processes, and applications. Adv Mater Technol. 2019;4(5):1800546.
Crossref - Pham N-D, Duong M-M, Le M-V, Hoang HA, Pham L-K-O. Preparation and characterization of antifungal colloidal copper nanoparticles and their antifungal activity against Fusarium oxysporum and Phytophthora capsici. Comptes Rendus Chimie. 2019;22(11-12):786-793.
Crossref - Ali S, Hameed S, Shahid M, Iqbal M, Lazarovits G, Imran A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol Res. 2020;232:126389.
Crossref - Shalal OS. In silico Characterization and Antifungal Activity of Curcuma Longa on DNA Topoisomerases of Candida Auris. Ann Rom Soc Cell Biol. 2021:860-875.
- Amalraj A, Pius A, Gopi S, Gopi S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives-A review. J Tradit Complement Med. 2017;7(2):205-233.
Crossref - Behura C, Ray P, Rhthi CC, Mishra RK, Ramachandraiah OS, Charyulu JK. Antifungal activity of essential oils of Curcuma longa against five rice pathogens in vitro. Journal of Essential Oil-Bearing Plants. 2000;3(2):79-84.
- Martinez-Correa HA, Paula JT, Kayano ACAV, et al. Composition and antimalarial activity of extracts of Curcuma longa L. obtained by a combination of extraction processes using supercritical CO2, ethanol and water as solvents. J Supercrit Fluids. 2017;119:122-129.
Crossref - Avanco GB, Ferreira FD, Bomfim NS, et al. Curcuma longa L. essential oil composition, antioxidant effect, and effect on Fusarium verticillioides and fumonisin production. Food Control. 2017;73:806-813.
Crossref - Prakash B, Kedia A, Mishra PK, Dubey NK. Plant essential oils as food preservatives to control moulds, mycotoxin contamination and oxidative deterioration of agri-food commodities-Potentials and challenges. Food Control. 2015;47:381-391.
Crossref - Subba R, Mathur P. Functional attributes of microbial and plant based biofungicides for the defense priming of crop plants. Theor Exp Plant Physiol. 2022:1-33.
Crossref - AL-Masoodi H, Hussein HJ, Al-Rubaye AF. Antifungal activity of the two medicinal plants (Curcuma longa L. and Boswellia carteri Birdwood) against Fusarium species isolated from maize seeds. Int J Pharm Res. 2020;12(3):408-414.
Crossref - Kadokawa J-I, Shimohigoshi R, yamashita K, Yamamoto K. Synthesis of chitin and chitosan stereoisomers by thermostable a-glucan phosphorylase-catalyzed enzymatic polymerization of a-D-glucosamine 1-phosphate. Organic & Biomolecular Chemistry. 2015;13(14):4336-4343.
Crossref - Al-Khafaji MA, Al-Sultany HH. Influence of chitosan on hematological and histopathological changes in mice infected with Brucella melitensis immunized with Rev-1 vaccine. Iraqi J Vet Sci. 2020;34(1):23-29.
Crossref - Lopez-Moya F, Lopez-Llorca LV. Omics for investigating chitosan as an antifungal and gene modulator. J Fungi. 2016;2(1):11.
Crossref - Liu R, Xu C, Zhang Q, Wang S, Fang W. Evolution of the chitin synthase gene family correlates with fungal morphogenesis and adaption to ecological niches. Sci Rep. 2017;7(1):44527.
Crossref - Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010;13(4):416-423.
Crossref - Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23(3):185-202.
Crossref - Fraschini R. Cytokinesis in eukaryotic cells: the furrow complexity at a glance. Cells. 2020;9(2):271.
Crossref - Lesage G, Shapiro J, Specht CA, et al. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8.
Crossref - Lipovsky A, Nitzan Y, Gedanken A, Lubart R. Antifungal activity of ZnO nanoparticles-the role of ROS mediated cell injury. Nanotechnology. 2011;22(10):105101.
Crossref - Gupta M, Tomar RS, Kaushik S, Mishra RK, Sharma D. Effective antimicrobial activity of green ZnO nano particles of Catharanthus roseus. Front Microbiol. 2018;9:2030.
Crossref - Degola F, Berni E, Dall’Asta C, et al. A multiplex RT-PCR approach to detect aflatoxigenic strains of Aspergillus flavus. J Appl Microbiol. 2007;103(2):409-417.
Crossref - Malla S, Raut CG, Sivakumar U, Malla G, Nishitha A. Effect of wheat grass extracts (Triticum aestivum) on wound healing related proteins (syndecan4 and tissue transglutaminase). World Journal of Pharmacy and Pharmaceutical Sciences. 2016;4(11):1040-1054.
- Keservani RK, Sharma AK, Kesharwani RK. Nutraceuticals and Dietary Supplements: Applications in Health Improvement and Disease Management. 2020: CRC Press.
Crossref - Chen C, Long L, Zhang F, et al. Antifungal activity, main active components and mechanism of Curcuma longa extract against Fusarium graminearum. PloS one. 2018;13(3):e0194284.
Crossref - Kong L-A, Yang J, Li G-T, et al. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathogens. 2012;8(2):e1002526.
Crossref - Salci TP. New targets for the development of antifungals targeting invasive candidiasis. Dissertation. State University of Maringa; 2017.
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.