ISSN: 0973-7510
E-ISSN: 2581-690X
The present study included the detection of some virulence factors of Aeromonas hydrophila under molecular level to clinical isolates were taken from patients suffering from diarrhea during the period from July (2017) to October (2017). Molecular detection of Hemolysin gene (ahh) was done for all isolates. The results showed that all isolates (100%) gave positive results for this virulence gene. the positive results were detected by the presence of (130) bp bands when compared with allelic ladder. The genomic DNA of the samples was extracted and bands were observed by performing agarose gel electrophoresis. When PCR was performed, results clearly indicate that all isolated organisms contained serine protease gene and all the amplified products produced a band at the level of (900 bp) when compared with the allelic ladder. Molecular detection of this gene was carried out by using a specific PCR primer were done by comparison with allelic ladder which gave a (309bp) It was found that (Aerolysin) gene present in (12)(75%) of the positive samples. Lip gene was also detected in A. hydrophila samples and found that all 16 samples (100%) gave positive results to this gene which gave molecular length (382) bp. Molecular study was carried out to show the sequence identity of cytotonic enterotoxins gene in Aeromonas spp. to that in A. hydrophila. Analysis of the A. hydrophila genome revealed a number of a putative virulence factors, including a gene that heat-labile cytotonic enterotoxin (alt). Our study showed that all (16) isolates (100%) gave positive results to this gene, which gave molecular length (442)bp. Molecular detection of cytotonic enterotoxins gene (ast) was done for (16) A. hydrophila isolates and the results showed that all isolates have this gene (100%). The positive results for (ast) virulence were detected by the presence of (331) bp band compared with allelic ladder.
Aeromonas hydrophila, Hemolysin gene (ahh) Aerolysin) gene, cytotonic enterotoxins gene (ast).
The genus Aeromonas is previously placed in the family, Vibrionaceae based on its phenotypic expression1, but they have recently been transferred from Vibrionaceae to their new family Aeromonadaceae according to Molecular genetics2. The genome of Aeromonas hydrophila strain ATCC 7966T is completely sequenced3.The genome is comprised of a single circular (4.744), (448) bp, chromosome with (61.5%) GC content4. The complete genome sequence of Aeromonas hydrophila reveals mechanisms contributing to virulence and metabolic condition that allow the organism to grow in a variety of environments and explains how Aeromonas hydrophila is able to survive in polluted or oxygen- poor environments and to colonize and cause illness in humans and other hosts. Thus, Aeromonas hydrophila genome sequence provides valuable insights into its ability to flourish in both aquatic and host environments3. Detection of A. hydrophila by amplification of virulence genes through PCR is rapid, sensitive and less time taking. Detection of A. hydrophila has been carried out by amplification of aerolysin gene, Hemolysin gene and lipase gene5. The PCR amplification of Hemolysin gene from A. hydrophila has been reported. The specific, rapid and sensitive PCR technique has been used to detect Hemolysin and aerolysin in Indian isolates of A. hydrophila (2). An extracellular lipase (EC) is produced by A. piscicola AH-3 (formerly A. hydrophila); however, the link between virulence and this gene is speculative in A. hydrophila6. Lipase is also an important extra-cellular virulence factor affecting several immune system functions through free fatty acid generated by lipolytic activity7, characterize other nine lateral flagellar genes for A. hydrophila. The cytotonic enterotoxins do not cause degeneration of crypts and villi of the small intestine-like cytotoxic enterotoxin8. Knock out mutations in either the alt or ast gene of A. hydrophila, and subsequent challenge of mice with these mutant strains, showed significantly reduced accumulation of fluid in the ligated ileal loop of the animal model, compared with that of wild type, indicating a distinct role of these factors in diarrhea9. In the isolate A. hydrophila B32, a novel serine protease (ser) was found that exhibits cytotoxic properties and is thermo stable, both of which hare characteristics that differentiate this protease from known A. hydrophila á- hemolysin and ß-hemolysin10.
The present study is carried out to detection the virulence genes associated with important virulence factors include: hemolysin (ahh), aerolysin (aer), Lipase (lip), Serine protease (ahp), thermo labile cytotonic enterotoxin (alt), thermo stable cytotonic enterotoxin (ast).
Samples collection and identification of A. hydrophila
According to previous study of11.
DNA extraction form bacterial culture
Genomic DNA was extracted from the Aeromonas hydrophila isolates according to instruction provided by manufacturer using Genomic DNA purification kit supplemented by (Geneaid, USA). The viewed using UV-trans illuminator:
- bacterial cells (in the sediment) was collected from swab samples that diluted in phosphate buffer solution and transferred to a 1.5 ml micro centrifuge tube then centrifuged in high speed centrifuge at 14000-16000 rpm for 1 minute then the supernatant discarded.
- Add 20 µl of proteinase k (make sure ddH2O was added ).incubate at 60ºC for 10 minutes and inverted every 3 minutes through incubation periods
- GB buffer (200µl) were added to each tube and mixed by shaking vigorously for 5 seconds. Then the tubes were incubated in water bath at 60ºC for 10 minutes and inverted every 3 minutes through incubation periods.Meanwhile, Elution buffer (200µl per sample) was pre-heated at 60°C (to be used in step 9 DNA Elution).
- Absolute ethanol (200 µl) were added to the clear lysate and immediately mixed by shaking vigorously, and then precipitates broke it up by pipetting.
- A GD column was placed in a 2 ml collection tube and transferred all of the mixture (including any precipitate) to the GD column. Then centrifuged at 14,000-16,000 rpm for 2 minutes. In addition, the 2 ml collection tube containing the flow-through were discarded and placed the GD column in a new 2 ml collection tube.
- W1 buffer (400µl) were added to the GD column, then centrifuge at 14,000-16,000 rpm for 30 seconds. The flow-through was discarded and placed the GD column back in the 2 ml collection tube.
- Wash Buffer (ethanol was added) 600µl were added to the GD column. Then centrifuged at 14,000-16,000 rpm for 30 seconds. The flow-through was discarded and placed the GD column back in the 2 ml collection tube.
- All the tubes were centrifuged again for 3 minutes at 14,000-16,000 rpm to dry the column matrix.
- The dried GD column was transferred to a clean 1.5 ml micro centrifuge tube and 150 µl of pre-heated elution buffer were added to the center of the column matrix.
- The tubes were let stand for at least 3 minutes to ensure the elution buffer was absorbed by the matrix. Then centrifuged at 14,000-16,000 rpm for 30 seconds to elute the purified DNA.
- The extracted DNA was stored at -16°C. Later on the extracted DNA that used for molecular identification of A. hydrophila and its virulence factors.
Primers Sequences
The primers sequences and PCR conditions that used in study are listed in Table (1).
Table (1):
the primers, sequences, and PCR conditions
Gene name |
Primer sequence (5´- 3´) |
Size of Bp |
Conditions |
References |
|---|---|---|---|---|
Aero |
F- ATGCTGCAGAAATGA ATAGAATAATTACCGC
R- ATGCAAGGCTTGCCCCATAA TCTCCCAGCGAT |
301 |
95/5min/30 cycle
95/2 min 55/1 min 72/1 min 72/7 min |
Xai et at., 2003 |
Hly |
F- CTATGAAAAAACTAAAAATAACTG
R- CAGTATAAG TGGGGAAATGGAAAG |
130 |
95/5min/30 cycle
95/2 min 55/1 min 72/1 min 72/7 min |
Xai et al, 2003 |
Ast |
F- TCTCCATGCTTCCCTTCCACT
R -GTGTAGGGATTGAAGAAGCCG |
331 |
95/5min/30 cycle
95/25Sec 55/30Sec 72/1 min 70/5min |
K.Sen et al., 2004 |
Alt |
F -TGACCCAGTCCTGGCACGGC
R -GGTGATCGATCACCACCAGC |
442 |
95/5min/30 cycle
95/25Sec 55/30Sec 72/1 min 70/5min |
K.Sen et al., 2004 |
Lipase (Lip) |
F- ATCTTCTCCGACTGGTTCGG
R- CCGTGCCAGGACTGGGTCTT |
382 |
95/5min/30 cycle
95/25Sec 55/30Sec 72/1 min 70/5min |
K.Sen et al., 2004 |
Ahp |
F- ATTGGATCCCTGCCTA
R- GCTAAGCTTGCATCCG |
911 |
94/5min/30 cycle
94/30Sec 56/30Sec 72/1 min 72/7min |
Contents of Reaction Mixture
Contents of Reaction Mixture
Amplification of DNA done in final volume of (25µ) containing the following as in Table (2).
Table (2):
Polymerase Chain Reaction Mixture
| No. | Mixture Contents | Volume (µl) |
|---|---|---|
| 1 | Master Mix | 12.5 |
| 2 | Forward Primer | 2.5 |
| 3 | Reverse Primer | 2.5 |
| 4 | Template DNA | 5 |
| 5 | Nuclase -Free Water | 2.5 |
| Total | 25 | |
Detection of Amplified Products by Agarose Gel Electrophoresis
Successful PCR amplification was confirmed by agarose gel/ electrophoresis by visualization against UV light (12). Agarose gel was prepared. Then the comb was fixed at one end of the tray for making wells used for loading DNA sample. The agarose was poured gently into the tray, and allowed to solidify at room temperature for 30 min. The comb was then removed gently from the tray. The tray was fixed in an electrophoresis chamber filled with TBE buffer that covered the surface of the gel, 5µl of DNA sample was transferred into each well of agarose gel, and in one well we put the 5µl DNA ladder. The electric current was allowed to pass at 70 volts for 50min. UV trans-illuminator was used 280 nm for the observation of DNA bands, and the gel was photographed using digital camera.
Detection of Virulence Factors Genes
A. hydrophila strains have produced a variety of biologically active extracellular products similar to the virulence factors of Enteropathogenic bacteria and these virulence factors associated with health effects in humans13. A. hydrophila produce virulence factors such as haemolysin, aerolysin, proteases, lipases DNAase and enterotoxins, which are important in the pathogenesis of human and fish disease14, 15, 16.
Detection of Hemolysin Gene (ahh)
Molecular detection of Hemolysin gene (ahh) was done for 16 isolates that previously detected as A. hydrophila. The results showed that all isolates (100%) gave positive results for this virulence gene. the positive results were detected by the presence of (130) bp bands when compared with allelic ladder as shown Figure (1). The present results agree with17, 18, 16 confirmed that A. hydrophila strains carried genes encoding hemolysin and the â-haemolytic A. hydrophila strains in fish and fish products available for human consumption throughout food stores represent a risk to the consumers health. According to19, these strains could be dangerous and are more likely to cause food poisoning in humans. Hemolysins are exotoxins and the lytic activities on red blood cells are reported to be important for nutrient acquisition or for causing anemia20. Hemolytic toxins as hemolysin and aerolysin released by A. hydrophila21 and play significant role in pathogenesis. Hemolysin genes did not appear to be distributed randomly among the different Aeromonas species. The hemolytic activity of A. hydrophila has been used as an indicator of enterotoxicity and may be responsible for outbreaks of diarrhea22. Nucleic acid amplification methods targeting virulence genes are used for detection of pathogenic bacteria and to differentiate pathogenic from non-pathogenic strains23, 24.
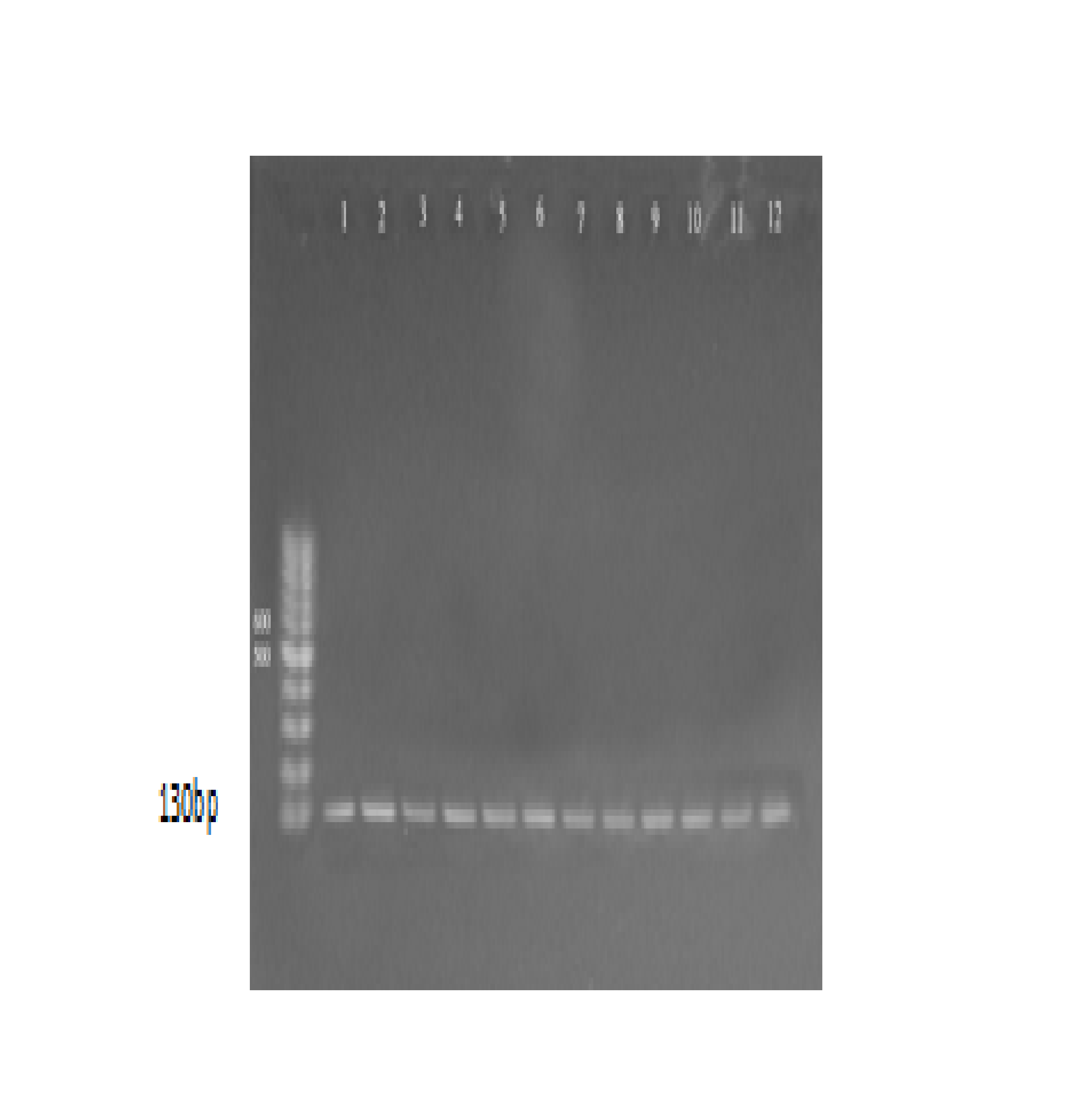
Fig. 1. agarose gel electrophoresis (1.5%) of RCR amplified of hly gene (130)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
Detection of Serine protease Gene (ahp)
Molecular studies of serine protease gene were done for all A. hydrophila samples by using specific PCR markers. Polymerase chain reaction is a sensitive and specific method for identification of virulence gene. Hence, for the direct detection of pathogenic Aeromonas species isolates, virulence determinants are used as a genetic marker. Thus, PCR method is used to detect the virulent gene encoding serine protease using specific primer PCR technique clearly identified the virulent gene as serine protease gene in Aeromonas hydrophila. The genomic DNA of the samples was extracted and bands were observed by performing agarose gel electrophoresis. When PCR was performed, results clearly indicate that all isolated organisms contained serine protease gene and all the amplified products produced a band at the level of (900 bp) when compared with the allelic ladder as showed in Figure (2)25 studied the serine protease and reported the PCR amplified band at (900 bp). The results of the present study also confirm the same26; all samples were give positive results 16 positive (100%) for serine protease gene with variable amplicon when it was compared with allelic ladder. Proteases produced by bacteria have been shown to cause tissue damage, aid invasiveness and establishment of infection by overcoming host defenses and provide nutrients for bacterial proliferation. It was reported that Aeromonas produced several kinds of protease. Studies of the proteases of Aeromonas have mainly been performed with A. hydrophila27. It has been suggested that proteolytic enzymes of human pathogen, Aeromonas hydrophila, play an important role in causing massive tissue damage in the host, which may facilitate establishment of infection28. Serine protease or serine endopeptidases are proteases (enzymes that cut peptide bonds in proteins) in which one of the amino acids at the active site is serine. Serine protease is known to activate toxins such as aerolysin and Glycerophospholipid Cholestrol acyl transferase (GCAT). This process is called quorum-sensing29. Glycerophospholipid cholesterol acyl transferase (GCAT) and Aerolysin both are activated by serine protease. The results also contrary with23 which stated that aerolysin / hemolysin and serine protease genes were present in all â – hemolytic strains supporting serine protease act as possibly important for the activation of the former gene. PCR detection may prove to be an important tool for the detection, identification, differentiation, and distribution of virulence markers. This method will give microbiologists a substitute way to understand pathogenicity in Aeromonas spp. and their distribution in different sources of diarrheal sample. The present report describes PCR method that detects serine protease genes in A. hydrophila by using a pair of primers for a serine protease gene the most known virulence gene of A. hydrophila.
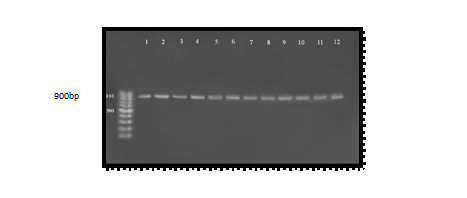
Fig. 2. Agarose gel electrophoresis (1.5%) of RCR amplified of ahp gene (900)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
Detection of aerolysin Gene (aerA)
Molecular detection of this gene was carried out by using a specific PCR primer were done by comparison with allelic ladder which gave a (309bp) It was found that (Aerolysin) gene present in (12)(75%) of the positive samples as shown in Figure (3). The results were closely related to those obtained by30 who mention that A. hydrophila have ability to produce aerolysin. Also our results agreement with31 who reported that the distribution of Aerolysin gene reach to (85.7%). Aerolysin was the factor most commonly described in A. hydrophila isolates, and previous studies have shown that two hemolytic toxins: HlyA and aerA contribute to the virulence of A. hydrophila. However, the inactivation of either aerA or hlyA alone reduced but did not totally cancel the hemolytic and cytotoxic activities of virulent strains of A. hydrophila .These activities were removed only when both the hlyA and aerA genes were inactivated. It is likely that both toxins act by pore-formation and the effects are likely to be synergistic32. Current study in agreement with other papers 33, 34. The presence of aerolysin is a strong indication of virulence in pathogenic isolates of Aeromonas spp35. The major hemolysin produced by aeromonads is called aerolysin, though it is known by several other names (cytotoxic enterotoxin, Asao toxin, and cholera toxin crossreactive cytolytic enterotoxin). Aerolysin is produced by some strains of A. hydrophila36. Aerolysin is a pore-forming toxin and is regarded, as the most important virulence factor in Aeromonas food poisoning, produced by some strains of A. hydrophila, is an extracellular, soluble, hydrophilic protein exhibiting both hemolytic and cytolytic properties37.
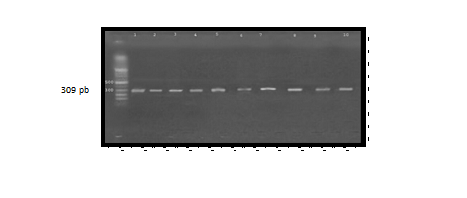
Fig. 3. Agarose gel electrophoresis (1.5%) of RCR amplified of aerA gene (309)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
Detection of Lipase Gene (lip)
Lipases may be important for bacterial nutrition and they may constitute virulence factors by interacting with human leukocytes or by affecting several immune system functions through free fatty acids generated by lipolytic activity34. Lip gene was also detected in A. hydrophila samples and found that all 16 samples (100%) gave positive results to this gene, which gave molecular length (382) bp. The amplicon was detected in gel electrophoresis and compared with allelic ladder as shown in Figure (4), our results agree with38 who mention that A. hydrophila have ability to produce lipase. The enzyme have essential role in pathogenicity and enhanced bacterial persist, the direct role of lipase to hydrolysis of phospholipids leading to depletion of structural lipids or generation of lytic reaction products both leading to loss of membrane integrity and cytotoxic. The lipases and hydro lipases were considered as important virulence factors in Aeromonas spp. because they alter the structure of the cytoplasmic membrane of the host and thus exacerbate its pathogenicity, especially if the aerolysin gene is present33. These genes are important extracellular factors for colonization of host tissues and their necrosis 39,40 demonstrated the potential of the association of elastase and lipase with the damage caused by aerolysin in cell cultures. The capacity of extracellular enzymes to cause lysis to “feed” the bacterial cells in proliferation is very important for Aeromonas spp39. The lipase gene was also described as an important virulence factor in Aeromonas spp. isolates from trout33,35 proposed that for A. hydrophila the presence of two genes (Lipase and aerolysin) might indicate their virulence in animals. These results confirm that this species has a larger matrix of virulence genes when compared with other species of clinical relevance41.
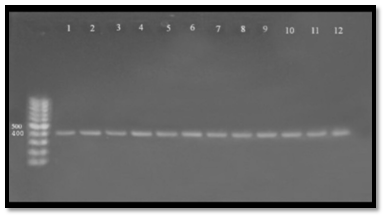
Fig. 4. Agarose gel electrophoresis (1.5%) of RCR amplified of lip gene (382)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
Detection of cytotonic enterotoxins Gene (alt)
Cytotonic enterotoxins may play significant roles in the pathology of Aeromonas gastrointestinal disease41. Molecular study was carried out to show the sequence identity of cytotonic enterotoxins gene in Aeromonas spp. to that in A. hydrophila. Analysis of the A. hydrophila genome revealed a number of a putative virulence factors, including a gene that heat-labile cytotonic enterotoxin (alt). Our study showed that all16 isolates (100%) gave positive results to this gene, which gave molecular length (442)bp as shown in Figure (5). Alt is a Heat-labile cytotonic enterotoxins exhibiting a size of 44 kDa single polypeptide that causes elevation of cyclic AMP and prostaglandin levels in Chinese hamster ovary (CHO) and intestinal epithelial cells. The Cytotonic enterotoxins (ast, alt) do not cause degeneration of crypts and villi of the small intestine-like cytotoxic enterotoxin8. Mutations in either the alt or ast gene of A. hydrophila, and subsequent challenge of mice with these mutant strains, showed significantly reduced accumulation of fluid in the ligated ileal loop of the animal model, compared with that of wild type, indicating a distinct role of these factors in diarrhea9. The alt gene however does not manifest any lipase activity43. The results from44 suggest that the product of both the alt and ast genes may act synergistically to induce severe diarrhea. Extracellular factor genes and virulence in A. hydrophila32. In a large survey of enterotoxin genes (act, alt and ast) in Aeromonas spp. Isolates from children with diarrhea, healthy controls and the environment, there was a significant association between the presence of both alt and ast genes amongst diarrheal children compared with controls44.
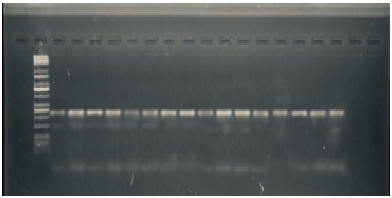
Fig. 5. Agarose gel electrophoresis (1.5%) of RCR amplified of alt gene (442)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
Detection of cytotonic enterotoxins Gene (ast)
Molecular detection of cytotonic enterotoxins gene (ast) was done for (16) A. hydrophila isolates and the results showed that all isolates have this gene (100%). The positive results for (ast) virulence were detected by the presence of (331) bp band compared with allelic ladder as shown in Figure (6). The presence of both alt and ast genes has been associated with sever watery diarrhea in A. hydrophila. However, the prevalence of these genes varies considerably depending on the source, species, geographical location and the number of isolates tested (41). The exotoxins are major virulence factors of Aeromonas spp. that include a cytotoxic heat-labile enterotoxin (act), a cytotonic heat-labile enterotoxin (alt) and a cytotonic heat-stable enterotoxin (ast) encoded by act, alt and ast gene, respectively (9, 45). In this study, all the isolates among Aeromonas spp. carried one or more enterotoxin genes. In the PCR assay, Aeromonas strains were found with different virulence gene combinations. The alt and ast genes showed the highest occurrence in A. hydrophila and all the strains carried these enterotoxin genes. The results are closely related to those results obtained by (45) who mention that (88%) of A. hydrophila carry the ast gene that have a role in diarrhea cases.

Fig. 6. Agarose gel electrophoresis (1.5%) of RCR amplified of ast gene (442)bp of A. hydrophila for (55)min at (70) volt; L: ladder (DNA marker). (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) Amplify of 16Sr RNA gene in isolates of A. hydrophila
- Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 2005; 242: 231-240.
- Yogananth, N., Bhakyaraj, R., Chantchuru, A., Anbalagan, T., Mullai Nila, K. Detection of virulence gene in Aeromonas hydrophila isolated from fish samples using PCR technique. Global. J. Biotechnol. Biochem. 2009; 4: 51–53.
- Daniel, H., Seshadri, R., Joseph, S. W., Chopra, A. K., Sha, J., Shaw, J Graf, J., Wu, M., Ren, Q., Rosovitz, M. J. and Heidelberg. J. F. Genome Sequence of Aeromonas hydrophila ATCC 7966T: Jack-Of-All-Trades. J. Bacteriol 2006.
- Vilches, S., Canals, R., Wilhems, M. ). Mesophilic Aeromonas UDP-glucose pyrophosphorylase (GaIU) mutants show two types of lipopolysaccharide structure and reduced virulence. Microbiology, 2007; 153: 2393-2404.
- Singh, V., Rathore, G., Kapoor, D., Mishra, B. N., Lakra, W. S. Detection of aerolysin gene in Aeromonas hydrophila isolated from fish and pond water. Indian. J. Microbiol. 2008; 48: 453–458.
- Anguita, J., RodriguezAparicio, L. B. and Naharro, G. Purification, gene cloning, amino acid sequence analysis, and expression of an extra cellular lipase from an Aeromonas hydrophila human isolate. Appl. Environ. Microbiol. 1993; 59: 2411–2417.
- Gavin, R., Rabaan, A. A., Merino, S., Toma´s, J. M., Gryllos, I., Shaw, J. G. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 2002; 43: 383–397.
- Sha J., Kozlova E. V. and Chopra A. K. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: Generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 2002; 60(4):1924-1935.
- Sha, J., Kozlova, E. V. and Chopra, A. K. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infection and Immunity. 2002; 70: 1924–1935.
- Esteve, C. and Birbeck, T. H. Secretion of heamolysin and proteases by Aeromonas hydrophila EO63: separation and characterization of the serine protease (caseinase) and the metallo protease (elastase). J. Appl. Microbiol. 2004; 96: 994–1001.
- Bunyan, I. A. and Obais, I. K. Molecular identification and phenotypic detection of some virulence factors among Aeromonas hydrophila isolated from diarrhea cases. J. Global Pharma Technol. 2018; 10(4-5):
- Sambrook, J. and Russel, D. W. Molecular Cloning: A Laboratory Manual 3rd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor 2001.
- Singh, V., Rathore, G., Kapoor, D., Mishra, B. N., Lakra, W. S. Detection of aerolysin gene in Aeromonas hydrophila isolated from fish and pond water. Indian. J. Microbiol. 2008; 48: 453–458.
- Castro-Escarpulli, G., Figueras, M. J., Aguilera-Arreola, M. G., Soler, L., Fernndez-Rendn, E., Aparicio, G. O., Guarro, J., Chacón, M. R. Characterization of Aeromonas spp. isolated from frozen fish intended for human consumption in Mexico. Int. J. Food. Microbiol. 2003; 84(1): 41-49.
- Chopra, A. K., J. W. Peterson, X.-J. Xu, D. H. Coppenhaver, and C. W. Houston. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 2014; 21: 357–377.
- Stratev, D. E., Gurova, A. I. Daskalov, H. Multiple PCR detection of hemolysin genes in â-hemolytic A. hydrophilia strains isolated from fish and fish products. Bulg. J. Agric. 2016; 22: 308-314.
- Wejdan, R., Taj Aldeen, F., Abeer, Al-Rubaiae, N. PCR Detection of Putative Hemolysin and Aerolysin Genes in An Aeromonas Hydrophila Isolates from Diarrhea in Babylon Province J. Natu. Scien. Research. 2014; 4(2): 224-318.
- Al-Fatlawy, H. N. K. and Al-Ammar, M. H. Study included the detection of A. hydrophila in some clinical cases in Iraq. International J. of Sci 2013; 2(21): 22113-6.
- Dar, G. H., Kamili, A. K., Chishti, M. Z., Dar, S. A., Tantry, T. A. Characterization of Aeromonas sobri isolated from fish Rohu collected from Polluted Pond. J. Bacterol. Parastiol. 2016; 7: 273.
- Griffiths, B. B. and McClain, O. The Role of Iron in the Growth and Hemolysin (Streptolysin S) Production in Streptococcus pyogenes. J. Basic Microbiology. 1988; 28(7): 427-436.
- Xia, C. Z., Ma, M., Habibur, R. and Zhi-Guang, W. PCR Cloning and Identification of the Haemolysin Gene of Aeromonas hydrophila from Freshwater Fishes in China. J.Aquaculture. 2003; 229(1): 45-53.
- Abbott S.L., Cheung W. K. W. and Janda, J. M. The Genus Aeromonas: Biochemical characteristics, Atypical Reactions, and phenotypic Identification Schemes. J. Clin. Microbiol., 2010; 41(6): 2348-2357.
- Chacon, M. R., Figueras, M. J., Castro-Escarpulli, G., L. Soler and J. Guarro. Distribution of Virulence Genes in Clinical and Environmental Isolates of Aeromonas spp. Antonie. Van. Leeuwenhoek. 2003; 84: 269-278.
- Sen, K. and Rodgers, M. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J. Appl. Microbiol. 2004; 97: 1077–1086.
- Li, J., Ni, X. D., Liu, Y. J. and Lu, C. P. Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebra fish. J. Appl. Microbiol. 2010; 110(3): 823-830.
- Martinez-Murcia, A. J., Benlloch, S. and Collins, M. D. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 1992; 42: 412–421.
- Gonza´lez-Serrano, C. J., Santos, J. A., Garcý´a-Lo´ pez, M. L. and Otero, A. Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhea case. J. Appl. Microbiol. 2002; 93: 414–419.
- Gunnlaugsd´ otter, B. and Gudmundsd´ ottir, B. K. Pathogenicity of atypical Aeromonas salmonicida in Atlantic salmon compared with protease production. J. Applied Microbiology. 1997; 83(5): 542–551.
- Gurcel, L., Iacovache, I., van der Goot, F. G. Aerolysin and related Aeromonas toxins. In The Comprehensive Source book of Bacterial Protein Toxins. London: Academic Press 2006.
- Yatanan, C. B., Adjehi, D., N’dédé, T. Djenil, B., Antoine, Y., Agathe, F. and Koffi, M. D. Molecular detection of hemolysin and aerolysin gene of Aeromonas spp. isolated from African tigrine frog (Hoplobatrachus occipitalis). Asian Pac. J. Trop. Dis. 2017; 7(1): 39-42.
- Coenye, T., Vandamme, P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 2003; 228: 45-49.
- Li, X., Yan, Z. and Xu, J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology. 2003; 149: 353-362.
- Nam, I. Y. and Joh, K. Rapid detection of virulence of Aeromonas isolated from a trout by hexaplex-PCR. J. Bacteriol. 2007; 45(4): 297-304.
- Nawaz, M., Khan, S. A., Khan, A. A., Sung, K., Tran, Q., Kerdahi, K. and Steele, R. Detection and characterization of virulence and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010; 27: 327-331.
- Heuzenroeder, M. W., Wong, C. Y. and Flower, RL. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS. Microbiol. Lett. 1999; 174: 131–136.
- Yousr, A. H., S. Napis, G. R. A., Rusul and Radu, R. Detection of Aerolysin and Hemolysin Genes in Aeromonas spp. Isolated from Environmental and Shellfish Sources by Polymerase Chain Reaction. ASEAN. Food. J. 2007; 14(2): 115-122.
- Qrmena, Q., M. Q., Reguea, J. and M. Tom. Studies of aerolysin promoters from different Aeromonas spp. Microb. Pathog. 2003; 35: 189-96.
- Samira, T. L., Oliveira, G., Veneroni, G. Mateus, M. C. Molecular characterization of virulence factors in Aeromonas hydrophila obtained from fish. Pesq. Vet. Bras. 2012; 32(8): 55-61.
- Cascon, A., Yugueros, J., Temprano, A., Sanchez, M., Hernanz, C., Luengo, J. M. and Naharro, G. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 2000; 68(6): 3233-3241.
- Song, T., Toma, C., Nakasone, N. and Iwanaga, M. Aerolysin is activated by metalloprotease in Aeromonas veronii biovar sobria. J. Med. Microbiol. 2004; 53: 477-482.
- Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 2007; 242: 231-240.
- Pablos, M., Remacha, M. A., Rodríguez-Calleja, J. M., Santos, J. A., Otero, A., García-López, M. L. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 2010; 29: 1163-1172.
- Chopra, A. K., J. W. Peterson, X.-J. Xu, D. H. Coppenhaver, and C. W. Houston. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog., 1996; 21: 357-377.
- Albert, M. J., Ansaruzzaman, M., Talukder, K. A., Chopra, A. K., Kühn, I., Rahman, M., Faruque, A. S. G., Islam, M. S., Sack, R. B. and Möllby, R. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol 2000; 38: 3785–3790.
- Kingombe, C. I. B., Aoust, Y., Huys, G. and Kwan, J. Multiplex PCR Method for Detection of Three Aeromonas Enterotoxin Genes. Appl. Enviro. Microbial. 2010; 76: 425-433.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


