ISSN: 0973-7510
E-ISSN: 2581-690X
Escherichia coli serotype O25b:H4 is involved in human urinary tract infections. In this study, we sequenced and analyzed E. coli O25b:H4 isolated from a patient suffering from recurring UTI infections in an intensive care unit at Hera General Hospital in Makkah, Saudi Arabia. We aimed to determine the virulence genes for pathogenesis and drug resistance of this isolate compared to other E. coli strains. We sequenced and analyzed the E. coli O25b:H4 Saudi strain clinical isolate using next generation sequencing. Using the ERGO genome analysis platform, we performed annotations and identified virulence and antibiotic resistance determinants of this clinical isolate. The E. coli O25b:H4 genome was assembled into four contigs representing a total chromosome size of 5.28 Mb, and three contigs were identified, including a 130.9 kb (virulence plasmid) contig bearing the bla-CTX gene and 32 kb and 29 kb contigs. In comparing this genome to other uropathogenic E. coli genomes, we identified unique drug resistance and pathogenicity factors. In this work, whole-genome sequencing and targeted comparative analysis of a clinical isolate of uropathogenic Escherichia coli O25b:H4 was performed. This strain encodes virulence genes linked with extraintestinal pathogenic E. coli (ExPEC) that are expressed constitutively in E. coli ST131. We identified the genes responsible for pathogenesis and drug resistance and performed comparative analyses of the virulence and antibiotic resistance determinants with those of other E. coli UPEC isolates. This is the first report of genome sequencing and analysis of a UPEC strain from Saudi Arabia.
Saudi, drug resistance, E.coli, UTI.
Uropathogenic Escherichia coli (UPEC) are ubiquitous and are involved in human urinary tract infections. Although several uropathogenic strains have been studied, the emergence of multidrug resistance with virulent phenotypes is a worldwide concern.1 The cause for the increased prevalence of multidrug resistance in E. coli is attributed to horizontal exchange of genetic material or mobile genomes.2 The E. coli O25b:H4 isolate is a uropathogenic pandemic clone primarily involved in the emergence of antimicrobial drug resistant community infections.3 The prevalence of virulent multidrug resistance genes of sequence type 131 (ST131) in E. coli O25b:H4 has been spreading widely due to its transferable plasmid. The emergence of clone ST131 O25b:H4 harboring extended-spectrum²-lactamase (ESBL) genes has been documented by several countries in Europe, Asia, and Middle East including Saudi Arabia.4,5 The sero group O25 is associated with ST131 and is linked to enterotoxigenic E. coli (ETEC)4. Whole genome sequencing studies have shown that E. coli ST131 strains encode virulence genes linked with extraintestinal pathogenic E. coli (ExPEC) and other virulence factor genes expressed constitutively in E. coli ST131. Therefore, the differences in virulence gene content may contribute to the variability in pathogenesis and host immunity.1 This work describes the whole genome sequencing and analysis of a UPEC strain that was isolated from a patient in the intensive care unit in a hospital in Saudi Arabia. We identified the functional role of this genome and performed targeted comparative analysis of virulence and antibiotic drug resistance determinants with other E. coli UPEC isolates.
DNA isolation
The E. coli O25b:H4 Saudi strain was isolated from a male patient admitted at Hera General Hospital in Makkah, Saudi Arabia. Bacteria was isolated from a single colony of E. coli O25b:H4 Saudi strain grown on 5% sheep blood agar and MacConkey agar, and genomic DNA was prepared using standard protocols from bacteria grown overnight in 5 ml LB broth at 37°C. The bacterial cells were centrifuged, and the cell pellet was used for genomic DNA extraction using the Qiamp DNA mini kit according to the manufacturer’s instructions (QiaGen, Valencia, CA, USA).
Genome sequencing and annotations
The E. coli O25b:H4 Saudi strain genome (WLH) was sequenced using multiple next-generation sequencing strategies. First, a random DNA library was constructed and paired-ends sequenced using the Illumina Mi Seq method, and they were assembled into 213 contigs using a CLC assembler. Second, the DNA was also sequenced from a library by PacBio SMRT cell. The sequence reads were assembled into 4 contigs using PacBio SMRT Analysis software (version 2.1.1, Pacific Biosciences, California, USA), and default filters removed reads <50 bases and less than 0.75 accuracy. The assembled PacBio verision was used for further bioinformatics and targeted comparative analysis. The open reading frames (ORFs) were identified using a combination of Glimmer (v 2.1), CRITICA and Prokpeg (a protein sequence similarity based ORF caller), as described in Kapatral et al.6 The reconciled predicted ORFs were integrated into the ERGO annotation environment for computing protein similarities and functional identification.7,8 The virulence and antibiotic resistance features were compared with other sequenced uropathogenic strains.
Genome accession
The genome sequences of the Escherichia coli 025b:H4 Saudi strain were deposited in GenBank with the accession numbers PRJNA316859 (SAMN04605558: Contig0001), PRJNA316859 (SAMN04605558: Contig0002), PRJNA316859 (SAMN04605558: Contig0003) and PRJNA316859 (SAMN04605558: Contig0004).
Phylogenetic analysis
We initially identified phylogenetic relationships between the hospital isolate and other pathogenic E. coli. Using three distinct phylogenetic marker DNA sequences, such as 16S rRNA, dnaA and gyrB sequences, against the Ribosomal Database Project (http:// rdp.cme.msu.edu) and NCBI database (http://blast.ncbi.nlm.nih.gov), our drug resistant hospital isolate was most similar to uropathogenic E. coli O25b:H4-ST131 str. EC958.
Genome analysis
To identify the virulence and drug resistant determinants, we sequenced the genome (WLH) and assembled it into four contigs. The total size of the genome was ~5.4 Mb with an average GC content of 50%. The genome features are given in (Table 1). It includes the chromosome of 5.2 Mb, three plasmids (13 kb, 3.2 kb, 2.9 kb). A total of 6,108 ORFs were identified in the chromosome, including rRNA and tRNA operons. The second contig was a plasmid containing the ORFs for the multidrug resistance bla-CTX gene, hemin receptor and nucleotidyl transferase protein. Using the ERGO annotation procedures described above, 70% of the ORFs were assigned with a functional annotation. Nearly 70% of ORFs functions belonged to COG categories,9 and 68% of ORFs had a pfam domain (10), signifying potential functions. A significant number of ORFs (49%) was identified as fusions proteins or frame shifts. Fusion proteins as composites (33%) consisting of two or more fused ORFs and ORFs representing individual components (34%) were identified.
Table (1):
Genome features of Escherichia coli O25b:H4 (WLH) Saudi strain.
Features |
Number |
Percent |
|---|---|---|
Total Contigs |
4 |
|
Total RNAs |
108 |
100 |
RNAs |
88 |
81 |
RNAs ribosomal |
12 |
11 |
Total ORFs |
6000 |
100 |
ORFs with assigned function |
4323 |
72 |
ORFs without assigned function |
1677 |
27 |
ORFs without assigned function with sims |
1645 |
27 |
ORFs in asserted pathways |
2584 |
43 |
ORFs not in asserted pathways |
3416 |
56 |
ORFs not in asserted pathways with assigned function |
1745 |
29 |
ORFs without assigned function with similarities |
1645 |
27 |
ORFs, in paralog clusters |
1864 |
31 |
ORFs in COGs |
4257 |
70 |
ORFs in Pfam domains |
4137 |
68 |
ORFs in operons/chromosomal |
5791 |
96 |
ORFs in possible fusion events |
2942 |
49 |
ORFs in possible fusion events as a composite |
1980 |
33 |
ORFs in possible fusion events as a component |
2057 |
34 |
Lipopolysaccharide (LPS)
LPS is a major component of the proteobacterial outer membrane and is a major virulence factor in all Gram-negative pathogens. Typically, Gram-negative bacterial LPS consists of three covalently linked moieties: the lipid A region, a conserved core oligosaccharide region and a serotype-specific O-antigen composition of polysaccharide side chains. ORFs for the synthesis of UDP-2,3-bis(3-hydroxymyristoyl)glucosamine biosynthesis, such as acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, RWLH04242, UDP-3-O-acyl-N-acetylglucosamine deacetylase, (RWLH04143) and UDP-3-O-(3-hydroxymyristoyl) glucosamine N-acyltransferase (RWLH04240) were identified. ORFs for lipid IVA biosynthesis viz., lipid-A-disaccharide synthase (RWLH04243) and tetra-acyldisaccharide 4′-kinase (RWLH05181) were identified in the same cluster. ORFs for the 2,3-diacylglucosamine-phosphate biosynthesis UDP-2 and 3-diacylglucosamine diphosphatase (RWLH04621) were also identified. Among the ORFs for inner core acylation, lipid A biosynthesis lauroyl acyltransferase (RWLH05312), lipid A biosynthesis lauroyl acyltransferase and lipid A biosynthesis (KDO) 2-(lauroyl)-lipid IVA acyltransferase (RWLH01427) were identified. An ORF for inner core biosynthesis related proteins, such as ADP-heptose-LPS heptosyltransferase III (RWLH02899), outer core polymerization O-antigen ligase (RWLH02887) and outer core translocation phospholipid-lipopolysaccharide ABC transporter (RWLH05180), was also found. Based on the structural organization of this operon, the WLH strain is contiguous with inner core “in frame” ORFs (E. coli strains, such as BW2952, DH10B, K-12, MG1655) or ORFs in frame with sequence differences (E. coli strains, such as 0111:H Str.1128, 55989, DE1, 11128, E2348/69, Sakai, UM026) compared to the opposite orientation in E. coli strains, such as APEC01, 536, ATCC8739, HS, IAI1 and UTI89.
Several ORFs were identified for outer core biosynthesis, such as alpha-L-glycero-D-manno-heptose alpha-1,3-glucosyltransferase (RWLH02898). Lipopolysaccharide core biosynthesis proteins, such as RfaS (RWLH02896), alpha-D-Glcp alpha-1,6-galactosyltransferase (RWLH02895), lipopolysaccharide 1,3-galactosyl-transferase (RWLH02894), lipopolysaccharide glucosyltransferase I (RWLH02893) and lipopolysaccharide core biosynthesis protein RfaZ (RWLH02891), were also identified. Two of the three inner core modification proteins, lipopolysaccharide core biosynthesis protein RfaP (RWLH02897) and lipopolysaccharide core biosynthesis protein RfaY (RWLH02892), were identified. Four ORFs for inner core biosynthesis components, such as (KDO)-lipid IV (A), 3-deoxy-D-manno-octulosonic acid transferase, 2.4.99.13/3-deoxy-D-manno-octulosonic-acid transferase/3-deoxy-D-manno-octulosonic-acid transferase (RWLH02901), lipopolysaccharide heptosyltransferase-1 (RWLH02886) and ADP-heptose-LPS heptosyltransferase II (RWLH02885), were identified.
Plasmid maintenance systems
As in other enteric bacteria, two ORFs belonging to the entericidin family operon ecnAB (RWLH03657 & RWLH03658) were identified. These proteins are involved in plasmid maintenance by post-segregation killing of cells (11).
Iron acquisition
Iron is a critical element for the survival of pathogenic bacteria inside the host, like in other pathogenic E. coli. We identified ORFs for siroheme biosynthesis, ORFs (RWLH03075, RWLH02591) for uroporphyrinogen-III C-methyltransferase and one ORF (RWLH02591) for precorrin-2 dehydrogenase and sirohydrochlorin ferrochelatase. Chelated iron uptake systems, such as Fe3 siderophore uptake outer membrane, siderophore receptor (RWLH00183), TonB (RWLH05614), TolQ (RWLH04835) and TolR (RWLH04836), were identified. An ORF for hemin receptor (RWLH01745, RWLH05731) for uptake of hemin; a homolog of yersiniabactin receptor for Fe3-yersiniabactin (RWLH00836); and a FhuE receptor precursor (RWLH05363) for Fe3-ferrioxiamineB, Fe3-hodotorulic acid and Fe3-coprogen were identified. Similarly, ferric aerobactin receptor (RWLH02061) and Fe3dicitrate transport FecA (RWLH00512, RWLH03835) were identified. A dedicated system for ferric enterobactin was identified, which included ferrienterobactin-binding protein FepB (RWLH04674), permease FepD (RWLH04672), FepG (RWLH04671), FepC (RWLH04670) and FepE (RWLH04669). Iron storage proteins, such as bacterioferritin (RWLH02554) and associated protein (RWLH02555), were also identified.
Adhesion
Several ORFs had adhesion/invasion functionality. An ORF similar to adhesion SefD (RWLH1073), invasion protein (RWLH00839, RWLH05418, RWLH04377, RWLH00840, RWLH00837, RWLH05574 and RWLH00838), in addition two ORFs (RWLH04280, RWLH00341) had similarities to NlpC/P60 family protein. One ORF for a lipoprotein antigen (RWLH02358) in other pathogenic bacteria, such as S. flexineri 5 Str.8401, E. coli APEC 01 and E. coli O157:H7, was identified Two ORFs with similarity to a polysaccharide involved in intercellular adhesion (RWLH00953, RWLH05282) and a polysaccharide deacetylase (RWLH04186) similar to other pathogenic enteric pathogens were identified. A polysaccharide as a biofilm operon pgaABCD involved in synthesis and export of 1-6-N-acetyl-D-glucosamine was identified. Upstream of this operon was a PGA transcriptional regulator. Similar to operons in other enteric pathogens, ORFs for secretin protein PgaA protein (RWLH05284), a specific deacetylase PgaB (RWLH05283), N-acetylglucosaminyltransferase PgaC (RWLH0582) and polymerization protein PgaD (RWLH05281), which are involved in the synthesis and export of biofilm, were identified.
Comparative secretion systems
Secretion of proteins in Gram-negative bacteria across inner and outer membrane is critical for pathogenesis. The comparative secretory systems found in the chromosome are summarized. Several secretory systems involved in protein translocation from the cytoplasm to periplasm or external milieu were identified in this genome as in other enterics. All the three stages of Sec-dependent pathway were present in the WLH genome, similar to other pathogenic enteric bacteria. This included an ORF for 4.5S RNA signal recognition particle (RWLH04543), signal recognition particle subunit SRP54 (RWLH01652), protein translocase subunit SecB (RWLH02874), chaperone protein DnaJ (RWLH04040), chaperone protein DnaK (RWLH00991, RWLH04038, RWLH04039), GrpE protein (RWLH01655), chaperonin GroES (RWLH03651) and chaperonin GroEL (RWLH03652). Similarly, all ORFs for the bacterial protein translocation pathway, such as secretion regulator SecM (RWLH04144), protein translocase subunit SecA (RWLH04145), SecD (RWLH04490), SecE (RWLH03278), SecF (RWLH04491), SecG (RWLH02385), SecY (RWLH02517) and YajC (RWLH04489), were identified. Other accessory proteins necessary for efficient translocation of proteins across the inner membrane, such as YidC protein (RWLH02988), trigger factor, ppiase (RWLH04521) and two ORFs for SecY stabilizing membrane protein (RWLH04883, RWLH05233), were identified. Among maturation and release proteins, one ORF for signal peptidase I (RWLH01606) and three ORFs for signal peptide peptidase SppA (RWLH00465, RWLH00648, RWLH05442) were identified. Similarly, a sec-independent protein system for the translocation of twin-arginine-motif containing proteins, requiring TatA protein (RWLH03131), TatB protein (RWLH03132), TatC protein (RWLH03133) and TatE protein (RWLH04709), was identified.
A critical secretion system necessary for the secretion of virulence proteins by the type II secretion (GSP) pathway was identified. ORFs for general secretion pathway protein A (RWLH02540); two ORFs for protein C (RWLH02153, RWLH02541, protein D (RWLH02152, RWLH02542), GPs protein (RWLH02151, RWLH02543, RWLH04152, and RWLH04153), GpsF protein (RWLH02150, RWLH02544, RWLH04151), Gps protein G (RWLH02149, RWLH02545), Gps protein H (RWLH02148), GPs protein I (RWLH02147, RWLH02547), Gps protein J (RWLH02146, RWLH02548), GPs protein K (RWLH02145, RWLH02549) Gps protein L (RWLH02550), Gps protein M (RWLH02143, RWLH02551) and Gps protein N; and two ORFs for type 4 prepilin peptidase (RWLH02552, RWLH02553) were identified, suggesting a functional GPS system. The ORF for Gps protein B was not identified. Virulence proteins secreted by type II secretion systems include alkaline phosphatase (RWLH04461), acid phosphatase (RWLH01760, RWLH03430, and RWLH03431, RWLH05245) and phosphatidyl-glycerophosphatase (RWLH00051, RWLH04500).
A secretion system for inserting specific proteins into the host membrane has been identified in several pathogenic bacteria (12). This includes proteins, such as translocase subunit SecB (RWLH02874), SecA (RWLH04145), translocase subunit YajC (RWLH04489), SecD (RWLH04490), SecF (RWLH04491), protein YidC (RWLH02988), SecE protein (RWLH03278), SecG protein (RWLH02385), SecY protein (RWLH02517), signal recognition protein FFH/SRP54 (RWLH01652), cell division protein FtsY (RWLH02695), signal peptidase I (RWLH01606) and ORF for lipoprotein signal peptidase (RWLH04054). This system is found in all pathogenic E. coli bacteria and appears to be functional, as well. Like in other E. coli UTI89 strains, we did not identify ORFs for a type III secretion system.
Motility
We identified both the flagellar and fimbrial types of motility that are typically found in enteric Gram-negative bacteria.
Flagellar motility
Two distinct flagellar systems were identified for motility and adhesion functionality, as in other pathogenic E. coli. The flagellar positive regulator protein transcriptional regulator LrhA (RWLH01335) and flagellar regulators, such as UmoB (RWLH02617) but not UmoA, UmoC or UmoD, were identified. ORFs for catabolite gene activator proteins (RWLH01141, RWLH02578) and DNA-binding protein H-NS (RWLH01673, RWLH05595), which are necessary for positive regulation of motility, were identified, as well. Both the ORFs for the flagella class-I pathway flagellar transcriptional activator FlhD (RWLH00607) and FlhC (RWLH00606), which turn on the class-II flagellar system, were identified.
Like in other enteric systems, flagellar class-II regulatory proteins, such as sigma factor flagellar operon FliA (RWLH00697, RWLH04330) and negative regulator of flagellin synthesis FlgM (RWLH04307, RWLH05330), were identified. ORFs for the flagella motor proteins pathway, including switch protein FliG (RWLH00714, RWLH04295), switch protein FliM (RWLH00720) and switch protein FliN (RWLH00721, RWLH04288), were identified. Among the hook-associated proteins, FlgE (RWLH04313, RWLH05336), flagellar hook-basal body complex protein FliE (RWLH00712, RWLH04293), hook-length control protein (RWLH00718, RWLH04328), and either hook-basal body complex protein FlhP or FlhO were found.
ORFs involved in the flagellar basal-body structure, such as basal-body rod modification protein FlgD (RWLH04311, RWLH05335), P ring formation protein FlgA (RWLH04308, RWLH05331), flagella synthesis protein FlgN (RWLH05329), assembly protein FliH (RWLH00715, RWLH04296), flagellar basal-body rod protein FlgB (RWLH04309, RWLH05333), rod protein FlgC (RWLH04310, RWLH05334), flagella rod protein FlgF (RWLH04314, RWLH05337) and rod protein FlgG (RWLH04315, RWLH05338), were identified. None of the other flagellar biosynthetic proteins that are found in other enteric bacteria, such as FlhB, FlhA, FlhF, FliP, FliQ and FliR, except for flagellar biosynthetic protein FliZ (RWLH00696) and murein transglycosylase (RWLH04013, RWLH05152), was identified in this genome. Among the flagella class-II structural proteins, ORFs for L-ring protein FlgH (RWLH05339), P-ring protein FlgI (RWLH04317, RWLH05340), flagellum-specific muramidase FlgJ (RWLH04318, RWLH05341), flagellar protein FliO (RWLH00722), flagellar basal body-associated protein FliL (RWLH00719), flagellar FliJ protein (RWLH00717), flagellum-specific ATP synthase (RWLH00716, RWLH04297) and assembly protein Flk (RWLH01371) were identified. However, ORFs for the flagellar synthesis regulator FleN and M-ring protein FliF were not found. The flagellar class-III structural proteins, such as flagellin (RWLH00698, RWLH043230), capping protein FliD (RWLH00700), hook-associated protein FlgL (RWLH04320, RWLH05343) and hook-associated protein FlgK (RWLH04319, RWLH05342), were identified. However, other structural proteins, such a flagellar protein FliS and FliT, were not identified in this genome.
Two sets of ORFs for the chemotaxis proteins MotB (RWLH00604, RWLH04332), MotA (RWLH00605, RWLH04331), protein-glutamate methylesterase (RWLH00597) and chemotaxis protein CheY (RWLH00596) were identified. None of the other chemotaxis proteins, such as CheA, CheD, CheC, CheW, CheV and CheZ, were identified. Interestingly, an ORF for aerotaxis receptor (RWLH02285), Tsr (RWLH03969), two ORFs for methyl-accepting chemotaxis protein Tar (RWLH00153, RWLH00601) and unknown methyl accepting chemotaxis protein (RWLH00599, RWLH00600) were identified in this genome.
Among the signal transduction systems, RcsB-RcsC containing three ORFs for the response regulator RcsB (RWLH01262), sensor protein RcsC (RWLH012630) and a specific sensor kinase YojN (RWLH01261), along with the QseB-QseC pathway consisting of the regulatory protein QseB (RWLH02221) and its cognate sensor protein QseC (RWLH02222), were identified. We conclude that both the lateral and peripheral flagellar systems are potentially functional in this genome.
Swarming motility
Two types of swarming motility pathways, namely type I and curli pilin, were identified in this genome.
Type I pili
All the ORFs necessary for type I pili synthesis were identified. These include an ORF (RWLH03604) for major pilin protein Fim A, three ORFs for type 1 regulatory protein FimB (RWLH04394, RWLH 03918, RWLH03921), outer membrane usher protein FimC (RWLH00229, RWLH03606, RWLH03925, RWLH04209, RWLH04210) and outer membrane usher protein FimD (RWLH00228, RWLH03526, RWLH03926). The type 1 fimbriae regulatory protein FimE (RWLH03922), FimF protein precursor (RWLH00227, RWLH01383, RWLH03927), FimG protein precursor (RWLH00226, RWLH03929), FimH protein precursor (RWLH00225, RWLH03930), fimbrin-like protein FimI (RWLH03924), type-1 fimbrial protein A chain precursor (RWLH00230, RWLH03923) and chaperone protein EcpD (RWLH03605, RWLH04198) were identified. However, other fimbrial proteins, such as V protein, W protein, Y protein, fimbriae Z protein or fimbrial protein FimX, were not identified in this genome.
Curli pilin
Curli pilin play a major pathogenic role by mediating adhesion and invasion into the host cell. The curli pilin proteins are also known to interact with host proteins, such as fibronectin, laminin, MHC class I proteins, TLR2 and fibrinogen, resulting in systemic infection. The curli pilin is encoded by two divergently placed operons: csgBAC (RWLH 05296, RWLH05297 and RWLH05298) and csgDEFG (RWLH05294, RWLH05293, RWLH0592, and RWLH05291). As in other entero-pathogenic bacteria, the role of the CsgC protein is unknown (13).
Multidrug resistance
Beta lactams
Resistance to antibiotics, such as such as penicillin, carbapenems and cephamycins, is due to the action of beta-lactamases. Three ORFs belonging to the beta-lactamase family, similar to non E. coli pathogens, have been identified: one ORF (RWLH05858) is similar to Klebsiella oxytoca, a second ORF (RWLH03661) is similar to pathogenic E. coli APEC 01 and the third (RWLH05853) is most similar to Pelobacter propionicus. Interestingly, upstream of the ORF (RWLH05853) is an ORF for aminoglycoside N6′-acetyltransferase, which is involved in aminoglycoside resistance; these two ORFs are flanked by an IS element that is located on the plasmid (contig 2: 139.5 kb) (Figure 1). It is interesting to note the divergence of two types of pathogenic E. coli. The genomes of WLH and E. coli 536 have identical organization at the ampicillin induction protein AmpE, unlike E. coli strains 0111:H, 11128, E2348/69 and EDL933 (Figure 2).
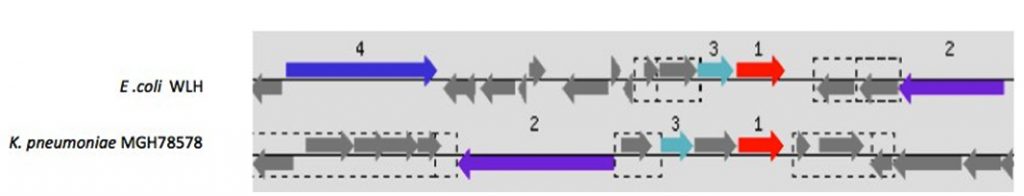 Fig. 1. The ORF for β-lactamase is predicted to have been acquired by transposition from other pathogens, such as Klebsiella spp.
Fig. 1. The ORF for β-lactamase is predicted to have been acquired by transposition from other pathogens, such as Klebsiella spp.1. β-lactamse,
2. Transposase,
3. Aminoglycoside N6’-acatyltransferase. Gray ORFs and boxed ORFs are parts of unknown transposons and IS elements, respectively
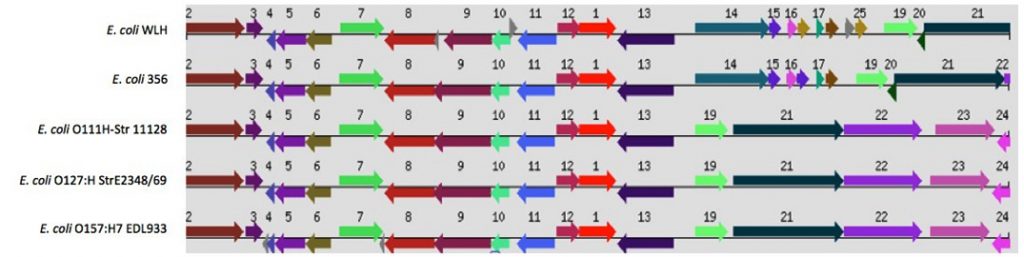
Fig. 2. The structural organization of Amp E induction protein is similar to E. coli 536; however, it is divergent from other pathogenic E. coli strains. 1. β-lactamase induction protein, AmpE, 2. Protein translocase subunit SecA, 3. 7,8-dihydro-8-oxoguanine-triphosphatase, 4. Hypothetical protein, 5. Hypothetical protein, 6. Dephospho-CoA kinase, 7. GMP reductase, 8. General secretion pathway protein F, 9. General secretion pathway protein E, 10. Type 4 major prepilin protein PilA, 11. Nicotinate-nucleotide pyrophosphorylase, 12. Anhydro-N-acetylmuramyl-tripeptide amidase, 13. Aromatic amino acid transport protein aroP, 14. Hypothetical protein, 15. Colicin E7 immunity protein, 16. Hypothetical protein, 17. Hypothetical protein, 18. Colicin E7 immunity protein, 19. Pyruvate dehydrogenase complex repressor, 20. Hypothetical protein, 21. Pyruvate dehydrogenase, 22. Dihydrolipoamide acetyl transferase, 23. Dihydrolipoamide dehydrogenase, 24. Hypothetical protein, 25. Colicin E7 immunity protein.
Tetracycline resistance
We identified two ORFs similar to the tetA family. One ORF similar to Salmonella spp and Acinetobacter spp was found in an IS element that was split into two ORFs, RWL05871 (792 bp) and RWLH05872 (447 bp). A second ORF (RWLH05709) (1272 bp), similar to the TetA protein of Acinetobacter spp, was identified in an IS element.
Chloramphenicol resistance
An ORF (RWLH02992) similar to the chloramphenicol export proton antiporter (multidrug efflux system protein MtdL) was identified, as in other pathogenic E. coli. These proteins are known to confer chloramphenicol resistance in several Gram-negative pathogens.
Multidrug resistance
Several ORFs that are known to confer multidrug resistance were identified: ORFs for multidrug resistance protein A YjcR (RWLH03476), EmrA (RWLH00117, RWLH01692) and multidrug resistance protein B YdiM (RWLH00380), as well as HrrA (RWLH03037), MdtD (RWLH00999), EmrB (RWLH01693), (YdiN) RWLH00381 (YdiN) and an unknown resistance protein (RWLH00118). One ORF was identified for each of the following multidrug resistance proteins: EmrD (RWLH02956), EmrK (RWLH01415), EmrY (RWLH01414) and Cmr (RLH04948). Two ORFs were identified for the Bcr family proteins (RWLH00071 & RWLH00348). Interestingly, three ORFs specifically for the Na+-driven multidrug efflux pump protein (RWLH00846) YeeO, (RWLH03350) DinF and (RWLH00352) MdtK were identified in the WLH genome.
Acriflavin resistance
Two ORFs with similarity to the enteric bacteria acriflavin-resistance proteins AcrA (RWLH04551) and AcrB (RWLH04550), which form a multidrug efflux system involved in protecting against hydrophobic inhibitors, such as antibiotics, detergents, disinfectants or dyes, were identified. In addition, other ORFs similar to the AcrA protein, such as MdtE (RWLH02763), AcrE protein (RWLH0283), MdtA protein (RWLH00996) as well as several ORFs similar to AcrB homologs, such as MdtB (RWLH00997), ArcD (RWLH01509), MdtC (RWLH00998), MdtF, (RWLH02764) and AcrF (RWLH0284), were present. As in other E. coli, the mdt operon containing ORFs for mdtABCD was identified.
Polymyxin resistance
These antibiotics possess a long hydrophobic tail and are known to be effective against Gram-negative bacteria. Resistance against these types of compounds is accomplished by expressing proteins, such as invasin. This protein is essential for invading the host cell membrane; the ORF for invasin was mutated at four sites, creating four non-functional ORFs. This frameshift is found in other pathogenic E. coli strains, such as 042, 536 and APEC 01.
We sequenced and analyzed a multidrug resistant uropathogenic E. coli isolated from a clinical setting from Saudi Arabia. Based on 16S RNA, gyrB and DNA sequence phylogenetic analysis, the strain was identified to be similar to uropathogenic E. coli O25b:H4 ST131. Like in other pathogenic E. coli, a number of drug resistant genetic determinants was identified along with other virulence, invasion, secretion and lysogenic phage genes. Interestingly, type III secretion systems necessary to deliver toxins into the host were not identified. Two sets of flagellar motility systems, lateral as well as peritrichous types, were identified. Similarly, two type I and curli pilin swarming systems appeared to be functional.
Multidrug resistant isolates of E. coli ST131 have been isolated from several countries in Asia and the Middle East; in Japan, approximately 21% of isolates were isolated between 2002-2003. A number of multidrug uropathogenic E. coli strains has been identified in Saudi Arabia (5); however, few strains have been fully sequenced. A huge genetic diversity was found within those isolates. The most notable diversity was among strains carrying CTX-M determinants (14). The prevalence of fluoroquinolone and ciprofloxacin resistant isolates (range 25-63%) was observed in various Japanese, Chinese and Philippines regions (15). These resistance markers were identified in this genome. In Lebanon, small hospitals reported E. coli with ESBL phenotypes from fecal samples (16). The exact epidemiology of E. coli ST131 clones in Saudi Arabia or other neighboring countries (e.g., Kuwait, UAE, Qatar, Bahrain, and Oman) remains unknown. However, the data is expected to be underestimated due to the lack of identification or reporting from these countries. Uropathogenic E. coli O25b: H4-ST131 strain EC958 is the biggest group of E. coli involved in extraintestinal infection. In 2005, E. coli O25b:H4-ST131 strain EC958 was isolated from a urine sample of a young girl diagnosed with UTI in the United Kingdom (17). The whole genome sequence of EC958 was studied, and it was found to contain the drug resistance gene blaCTX-M15 and multiple virulence factors involved with UPEC, including genes encoding autotransporter proteins (PicU, UpaH, UpaG and Ag43), adhesins (curli, type 1 fimbriae and a fimbrial adhesin) and siderophore biosynthesis genes (enterobactin, aerobactin and yersiniabactin). We found similar genes in our study, consistent with that by Totsika et al. (17).
A major dissemination of E. coli plasmid-mediated extended-spectrum ß-lactamases (ESBLs) producers has become evident worldwide similar to Acinetobacter baumannii which is rapidly emerging pathogen globally including Saudi Arabia (18). These bacterial strains, in addition to being resistant to ß-lactamases, are also resistant to aminoglycosides and fluoroquinolones. E. coli O25b:H4 ST131 is known for its CTX-M-15 production and spreading in both in-patients and out-patients globally (19). It harbors multiple drug resistant determinants on its plasmids in addition to virulence genes (20, 21). Consistent with previous studies, this study presents evidence of other drug resistant genes, such as tetracycline, chloramphenicol, aminoglycoside and acriflavins, and ESBLs determinants from this hospital isolate highlight its international dissemination (22). Consistent with our study, the gene acquisition by uropathogenic E.coli may enhance urinary tract infections and may aid in evading the host immune response (23).
ACKNOWLEDGMENTS
We acknowledge the national center for biotechnology and the Life Science & Environment Research Institute at KACST for facilitating and supporting the work of this project.
- Petty NK, Ben Zakour NL, Stanton-Cook M, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proceedings of the National Academy of Sciences 2014; 111:5694-9.
- Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Molecular microbiology 2006; 60:1136-51.
- Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 2011; 66:1-14.
- Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M ²-lactamases: the worldwide emergence of clone ST131 O25: H4. International journal of antimicrobial agents 2010; 35:316-21.
- Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. Journal of Antimicrobial Chemotherapy 2015:dkv188.
- Kapatral V, Anderson I, Ivanova N, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 2002; 184:2005-18.
- Overbeek R, Larsen N, Walunas T, et al. The ERGO genome analysis and discovery system. Nucleic Acids Res 2003; 31:164-71.
- Overbeek R, Begley T, Butler RM, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 2005; 33:5691-702.
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic acids research 2000; 28:33-6.
- Tatusov RL, Natale DA, Garkavtsev IV, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Research 2001; 29:22-8.
- Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. Journal of molecular biology 1998; 280:583-96.
- Jeeves M, Knowles TJ. A novel pathway for outer membrane protein biogenesis in Gram negative bacteria. Molecular microbiology 2015; 97:607-11.
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol 2006; 60:131-47.
- Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 2009; 63:72-9.
- Tian G-B, Garcia J, Adams-Haduch JM, et al. CTX-M as the predominant extended-spectrum ²-lactamases among Enterobacteriaceae in Manila, Philippines. Journal of antimicrobial chemotherapy 2010:dkp480.
- Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. Journal of Antimicrobial Chemotherapy 2008; 61:273-81.
- Totsika M, Beatson SA, Sarkar S, et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. 2011.
- Alyamani EJ, Khiyami MA, Booq RY, Alnafjan BM, Altammami MA, Bahwerth FS. Molecular characterization of extended-spectrum beta-lactamases (ESBLs) produced by clinical isolates of Acinetobacter baumannii in Saudi Arabia. Annals of clinical microbiology and antimicrobials 2015; 14:1.
- Vimont S, Boyd A, Bleibtreu A, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 2012; 7:e46547.
- Johnson JR, Johnston B, Clabots C, et al. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients. Antimicrobial agents and chemotherapy 2010; 54:546-50.
- Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clinical infectious diseases 2010; 51:286-94.
- Johnson JR, Anderson JT, Clabots C, Johnston B, Cooperstock M. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr Infect Dis J 2010; 29:473-5.
- Welch RA, Burland V, Plunkett G, 3rd, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 2002; 99:17020-4.
© The Author(s) 2016. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


