ISSN: 0973-7510
E-ISSN: 2581-690X
The manipulation of keratinolytic determinants provides an avenue for enhancing keratinase output, thereby facilitating the conversion of wasted chicken feathers into valuable products. Thus, this study investigated the keratinolytic determinants expressed by Chryseobacterium proteolyticum during the degradation of chicken feathers. The bacterial genome was sequenced using the Ion GeneStudio™ S5 Prime System, and SPAdes was used to assemble the raw sequences, while Prokka was used to annotate the obtained sequences. The keratinolytic genes were amplified by polymerase chain reaction (PCR). A total of 4.84 Mb bases were generated from the bacterial genome, which yielded thirty-seven contigs ranging between 165 bp and 1242714 bp when assembled by SPAdes. Prokka annotated 4798 genes, of which thirty-eight were identified as keratinolytic enzyme-coding genes. The resA coding for thiol-disulfide oxidoreductase was the most occurring keratinolytic determinant with a proportion of 26.3% (10/38), followed by dap coding for D-aminopeptidase (13.2%; 5/38). PCR revealed that resA exhibited a band size of 646 bp. The successful amplification of the keratinolytic determinants from this potent keratin degrader, C. proteolyticum, confirms its dexterity for keratinolysis and may pave the way for genetic engineering to enhance keratinase output.
Keratinase, Chicken Feathers, Disulfide Reductase, Aminopeptidases, Chryseobacterium proteolyticum
The increased consumption of chickens has resulted in the generation of millions of tons of chicken feathers annually, a waste with no efficient recycling strategies to manage the waste biomass.1 If handled poorly, poultry waste may pose a hazard to the environment and humans. Chicken feathers are insoluble and resistant to degradation due to the presence of a high amount of fibrous protein – keratin (90%). Keratin is structurally stabilized by the inherently interconnected disulfide, hydrogen, and hydrophobic bonds, and thus is resilient to decomposition under natural environmental conditions.2 Numerous waste management methods have been devised to curb chicken feather’s environmental accumulation. Typically, chicken feathers are disposed of through landfilling, and this disposal approach could promote the growth of pathogenic microbes, the release of offensive odours within the local vicinity and the eutrophication of waterbodies by the leachate.3 Incineration increases air pollution and contributes to the rising carbon footprint.3,4
Considering the keratin content in chicken feathers, many approaches have been explored to transform this waste into useful products with application potential in many sectors of the economy, which include the detergent, agro-sector, and pharmaceutical industries.5-7 The approaches that have been applied for the valorisation of the chicken feathers include high-pressure and temperature heating, mechanical crushing and chemical treatment of the biomass to generate protein hydrolysates for potential use in other sectors, such as animal feed and organic fertiliser. These processing technologies geared towards adding value to chicken feather biomass often damage the heat-sensitive amino acids, produce indigestible and low-quality products, consume energy, and introduce hazardous chemicals to the environment.8 The challenges orchestrated by traditional techniques necessitate the exploration of different ecological niches for microbial sources with the potential to express robust metabolic enzyme cocktails for valorising poultry feathers into high-value products.
Microbial keratinolytic enzymes, including disulfide reductases, carboxypeptidases, and aminopeptidases, are enzyme cocktails that display dexterity in dismembering keratinous biomass into amino acids, functional peptides, and non-protein nitrogenous compounds.9,10 The disulphide reductases are responsible for hydrolyzing densely populated cysteine disulfide bonds, and they include membrane-associated extra-cytoplasmic disulfide reductase (ResA)11 and Cytoplasmic disulfide reductase/Thioredoxin reductases (trxA and trxB).12 Similarly, aminopeptidases such as dipeptidyl aminopeptidase BIII (dapb-3) and aminopeptidase C (pepC) have all been associated with the bioconversion of chicken feathers to high-value products, post the breakage of disulfide bonds.13,14
Microbial keratinase-directed chicken feather degradation is a cost-effective and eco-friendly recycling method that is currently gaining traction in the bioindustry space. That, therefore, underscores the need to explore microbial keratinolytic enzyme-encoding genes to enhance keratinase yield for efficient bioconversion of chicken feathers into high-value products. The feather-degrading C. proteolyticum used for this study was previously isolated and characterized for its keratinolytic potential.15 Therefore, this study was implemented to sequence the genome of C. proteolyticum, identify the secondary metabolite determinants encoding for keratinolysis of chicken feathers and amplify the identified genes for enhancing keratinolytic activity. The identified keratinolytic enzyme-encoded genes have the potential to be amplified and used in genetic engineering to improve keratinase production. The sequenced genome was annotated to explore the keratinolytic determinants responsible for the keratinolytic activity of C. proteolyticum. The putative keratinolytic enzyme-encoding genes identified were successfully amplified.
Reagents
Media constituents were purchased from Merck Life Science Limited (Johannesburg, South Africa) and Lasec (Pty) Ltd. (Port Elizabeth, South Africa). Keratin Azure was obtained from Sigma-Aldrich (St. Louis, MO, USA). All the reagents used in the polymerase chain reaction (PCR), including primers, master mix, and nuclease-free water, were purchased from Inqaba Biotechnical Industries (Pty) Ltd. (Pretoria, South Africa); meanwhile, reagents used in gel electrophoresis were purchased from Lasec (Pty) Ltd. (Port Elizabeth, South Africa).
Feather keratin substrate preparation
Chicken feathers were collected from a local poultry farm in Middledrift, Raymond Mhlaba Municipality, South Africa. The chicken feathers were exhaustively washed with water, air-dried, and oven-dried at 60 °C for 48 h. The dry biomass was milled into powdered chicken feathers (PCF) using a 2 mm mesh-fitted pulverizer. The chicken feathers were kept in an air-tight container for subsequent use as a keratinous substrate.
Keratinolytic strain and culture media
The previously established keratinolytic C. proteolyticum identified through the 16S ribosomal RNA gene sequencing15 and maintained on basal salt feather-agar slants was utilized for this study. The bacterial strain was inoculated on basal salt medium (K2HPO4; 0.3%, KH2PO4; 0.4%, MgCl2; 0.2%, CaCl2; 0.22%) with chicken feather; 10%, and incubated at 30 °C for 48 h.16 This passaging process was repeated 4 times to reacclimate the bacterial strain to utilise keratinous feathers as the only source of carbon and nitrogen.
Keratinase production and enzyme assay
Keratinase production was carried out following the method previously reported.17 Concisely, a sterile basal salt media containing powdered chicken feather (PCF), 10 g/L as the only carbon and nitrogen source, was inoculated with a freshly prepared bacterial suspension (2%, v/v). The culture flasks were incubated for 96 h at 30 °C and 130 rpm (Labotec IncoShake (Pty) Ltd, Midrand, Gauteng, South Africa). Subsequently, the culture medium was filtered using a 0.45 µm pore filter and spun at 15,000x g in a centrifuge (Lasec SA (Pty) Ltd., Port Elizabeth, South Africa) for 10 min. The cell-free filtrate was used as a crude enzyme for keratinase assay, following the method described by Jaouadi et al.18 with some modifications. The keratinase assay reaction mixture was prepared by coupling 500 µL of crude enzymes with 500 µL of keratin azure solution (pH 7.5). The reaction mixture was incubated at 40 °C for 1 h with continuous shaking at 200 rpm. After the incubation, the reaction was terminated by placing the assay tubes on ice for 10 min. The keratinolytic activity was determined by measuring the amount of free azo dye released in the solution at 595 nm using a SYNERGYMx 96-well microplate reader (BioTek Instrument Inc., Winooski, VT, USA).
Whole genome sequencing (WGS) of the bacterial isolate
The genome of the C. proteolyticum was sequenced using Ion GeneStudio™ S5 Prime System (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, the genomic DNA (gDNA) of C. proteolyticum was isolated using the “salting out” method previously described by Sunnucks and Hales19 and purified using the magnetic beads method. The gDNA was quantified by the Qubit™ 4 Fluorometer using the Qubit™ 1X dsDNA High Sensitivity (HS) Assay Kit (Thermo Fisher Scientific), following the manufacturer’s protocol. The purity of the quantified gDNA was evaluated using NanoDrop® ND-1000 (Thermo Fisher Scientific). The genomic quality score (GQS) was determined on the LabChip GX Touch 24 Nucleic Acid Analyzer (PerkinElmer, Waltham, MA, USA) according to the manufacturer’s protocol. The gDNA sample was subjected to library preparation using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The Ion Library TaqMan™ Quantitation Kit quantified the prepared library by following the manufacturer’s protocol. The StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) was used to amplify the library using quantitative PCR to determine the molarity of the library and respective barcode. Subsequently, the amplified library was diluted to 40 pM and combined in equimolar amounts for template preparation using the Ion 510™, Ion 520™, and Ion 530™ Chef Kit (Thermo Fisher Scientific) and loaded on the Ion GeneStudio™ S5 Prime System for sequencing.
Sequence analysis and gene annotation
The obtained sequences from the Ion GeneStudio™ S5 Prime System were assembled using SPAdes assembler (Version 3.13.0)20 on the Stellenbosch University HPC (Western Cape, South Africa) and evaluated using QUAST V4.1.21 Genome annotation was performed using Prokka 1.14.6.22
The determination of the genetic bases for the expression of keratinolytic enzymes from C. proteolyticum
The primer basic local alignment search tool (BLAST) in the National Centre for Biotechnology Information (NCBI) (tool available publicly at http://www.ncbi.nlm.nih.gov/tools/primer-blast) was used to design primers for the target keratinolytic determinants from Prokka-annotated sequences. The PCR reaction consisted of 12.5 µL of Mastermix, 5 µL of DNA template, 1 µL of each primer, and 5.5 µL of nuclease-free water. PCR was executed in a Thermocycler (Bio-Rad Laboratories (Pty) Ltd, South Africa) programmed at 94 °C for 5 min (one cycle), followed by 30 cycles of 94 °C for 30 s, 50 °C for 1 min, 72 °C for 1 min, and one cycle of 72 °C for 10 min. The amplicons were loaded on 1.5% agarose gel and electrophoresed to confirm positive amplification of the test genes. The UV transilluminator (United Scientific (Pty) Ltd.) was used to view the DNA bands on the ethidium bromide-stained agarose gel.
Multiple-sequence alignment
The protein sequences similar to the successfully amplified target genes were searched on the protein database using the coding amino acid sequences retrieved from Prokka 1.14.6, and multiple sequence alignment was done to compare the sequences.
Determination of the physicochemical properties of the identified keratinolytic enzyme
The ExPASy-ProtParam tool was used to determine the molecular weight, amino acid composition, instability index, and theoretical isoelectric point (pI), as reported by Gasteiger and colleagues.23
Evaluation of keratinase production and activity
Revalidation of the keratinolytic potential of C. proteolyticum used for this study showed that the isolate secreted a significant amount of enzyme, which subsequently mediated the degradation of keratinous feather biomass in the fermentation medium, with keratinolytic activity and thiol concentration of 706.36 U/mL and 123.68 µM, respectively.
Whole genome sequencing, assembly and annotation
The Ion GeneStudio™ S5 Prime System generated 2,484,843 nucleotide base sequence reads from the C. proteolyticum genome length of 804,134,986, and SPAdes generated thirty-seven contigs. The maximum contig had a length of 1242714 bp, while the minimum contig consisted of 134 bp. The sequenced whole genome was uploaded on NCBI GenBank, and the accession number JBJLWK000000000 was generated. The Prokka annotation revealed that the bacterial genome consisted of 4733 coding sequences (cds), six rRNA, fifty-eight tRNA, and one tmRNA. Additionally, the annotation revealed that the keratinolytic C. proteolyticum under investigation possessed 4798 genes, where thirty-eight of these genes were coding for enzymes associated with keratinolysis, which included disulphide reductases, aminopeptidases, dipeptidyl peptidases, thioredoxin reductases, dipeptidases, and dipeptidyl carboxypeptidase. The various keratinolytic genes from C. proteolyticum, gene products, gene locus tag, and EC number are presented in Table.
Table:
Identified keratinolytic enzyme-encoding genes from the genome of C. proteolyticum
No. |
Gene |
Gene Locus Tag |
EC Number |
Clusters of Orthologous Genes (COG) |
Gene product |
|---|---|---|---|---|---|
1 |
pepE |
MLPAAHBK_00960 |
3.4.13.21 |
COG3340 |
Peptidase E |
2 |
pepC |
MLPAAHBK_01375 |
3.4.22.40 |
COG3579 |
Aminopeptidase C |
3 |
resA_1 |
MLPAAHBK_01466 |
– |
– |
Thiol-disulfide oxidoreductase |
4 |
sppA |
MLPAAHBK_01967 |
3.4.21.- |
COG0616 |
Protease 4 |
5 |
htpX |
MLPAAHBK_02208 |
3.4.24.- |
– |
Protease HtpX |
6 |
ptrB |
MLPAAHBK_02848 |
3.4.21.83 |
COG1770 |
Protease 2 |
7 |
trxA |
MLPAAHBK_03368 |
– |
– |
Thioredoxin A |
8 |
trxB |
MLPAAHBK_04313 |
1.8.1.9 |
COG0492 |
Thioredoxin reductase |
9 |
dapb1 |
MLPAAHBK_03502 |
3.4.14.- |
– |
Dipeptidyl aminopeptidase BI |
10 |
dapb3 |
MLPAAHBK_01060 |
3.4.14.- |
– |
Dipeptidyl aminopeptidase BIII |
11 |
pepS |
MLPAAHBK_00223 |
3.4.11.24 |
COG2234 |
Aminopeptidase S |
12 |
pepP |
MLPAAHBK_00699 |
3.4.11.9 |
COG0006 |
Xaa-Pro aminopeptidase |
13 |
cysH |
MLPAAHBK_00709 |
1.8.4.10 |
– |
Thioredoxin-dependent 5′-adenylylsulfate reductase |
14 |
cysJ_1 |
MLPAAHBK_00715 |
1.8.1.2 |
COG0369 |
Sulfite reductase [NADPH] flavoprotein alpha- component |
15 |
cysJ_2 |
MLPAAHBK_00716 |
1.8.1.2 |
COG0369 |
Sulfite reductase [NADPH] flavoprotein alpha- component |
16 |
cysI |
MLPAAHBK_00717 |
1.8.1.2 |
COG0155 |
Sulfite reductase [NADPH] hemoprotein beta-component |
17 |
dpp5 |
MLPAAHBK_01391 |
3.4.14.- |
COG1506 |
Dipeptidyl-peptidase 5 |
18 |
dpp7 |
MLPAAHBK_02697 |
3.4.14.- |
– |
Dipeptidyl-peptidase 7 |
19 |
pepQ |
MLPAAHBK_01509 |
3.4.13.9 |
– |
Xaa-Pro dipeptidase |
20 |
dap_1 |
MLPAAHBK_01627 |
3.4.11.19 |
– |
D-aminopeptidase |
21 |
dap_2 |
MLPAAHBK_01852 |
3.4.11.19 |
– |
D-aminopeptidase |
22 |
dap_3 |
MLPAAHBK_01862 |
3.4.11.19 |
– |
D-aminopeptidase |
23 |
dap_4 |
MLPAAHBK_02277 |
3.4.11.19 |
– |
D-aminopeptidase |
24 |
dap_5 |
MLPAAHBK_04392 |
3.4.11.19 |
– |
D-aminopeptidase |
25 |
resA_2 |
MLPAAHBK_01665 |
– |
– |
Thiol-disulfide oxidoreductase |
26 |
resA_3 |
MLPAAHBK_01869 |
– |
– |
Thiol-disulfide oxidoreductase |
27 |
resA_4 |
MLPAAHBK_01991 |
– |
– |
Thiol-disulfide oxidoreductase |
28 |
resA_5 |
MLPAAHBK_02098 |
– |
COG0526 |
Thiol-disulfide oxidoreductase |
29 |
resA_6 |
MLPAAHBK_02246 |
– |
– |
Thiol-disulfide oxidoreductase |
30 |
resA_7 |
MLPAAHBK_02658 |
– |
– |
Thiol-disulfide oxidoreductase |
31 |
resA_8 |
MLPAAHBK_02659 |
– |
– |
Thiol-disulfide oxidoreductase |
32 |
resA_9 |
MLPAAHBK_03252 |
– |
– |
Thiol-disulfide oxidoreductase |
33 |
resA_10 |
MLPAAHBK_04551 |
– |
– |
Thiol-disulfide oxidoreductase |
34 |
lon2 |
MLPAAHBK_02324 |
3.4.21.53 |
COG0466 |
Lon protease 2 |
35 |
pepT_1 |
MLPAAHBK_02676 |
3.4.11.4 |
COG2195 |
Peptidase T |
36 |
pepT_2 |
MLPAAHBK_02677 |
3.4.11.4 |
COG2195 |
Peptidase T |
37 |
dcp_1 |
MLPAAHBK_03703 |
3.4.15.5 |
COG0339 |
Dipeptidyl carboxypeptidase |
38 |
dcp_2 |
MLPAAHBK_03704 |
3.4.15.5 |
COG0339 |
Dipeptidyl carboxypeptidase |
The predominant keratinolytic enzyme-encoding gene that was observed from the C. proteolyticum genome was the thiol-disulfide oxidoreductase encoding gene (resA) with a proportion of 26.3% (10/38), followed by a D-aminopeptidase encoding gene (dap) with a proportion of 13.2% (5/38). While other keratinolytic-associated enzymes encoding genes such as dipeptidyl aminopeptidase encoding gene (dapb), dipeptidyl-peptidase encoding gene (dpp), peptidase T encoding gene (pepT), dipeptidyl carboxypeptidase encoding gene (dcp), and sulfite reductase [NADPH] flavoprotein alpha-component (cysJ) were identified in a proportion of 5.3% (2/38). While the genes coding for peptidase (pepE), aminopeptidase C (pepC), aminopeptidase S (pepS), Xaa-Pro aminopeptidase (pepP), Xaa-Pro dipeptidase (pepQ), protease 4 (spa), protease HtpX (htpX), protease 2 (ptrB), thioredoxin A (trxA), thioredoxin reductase (trxB), thioredoxin-dependent 5′-adenylylsulfate reductase (cysH), sulfite reductase [NADPH] hemoprotein beta-component (cysI), and lon protease 2 (lon2) were identified in a proportion of 2.6% (1/38). The high prevalence of the thiol-disulfide oxidoreductase encoding gene highlights the robust sulfolytic system of C. proteolyticum biosynthetic machinery and its involvement in the reduction of keratin’s disulfide bonds.
Multiple amino acid sequence alignment of the keratinolytic enzymes
Interestingly, about eight (80%) of the amplified keratinolytic enzymes (pepE, pepC, resA, trxA, trxB, cysH, dapb3, and dcp_1) displayed 99% similarity with the proteolytic enzymes from Chryseobacterium daecheongense strain PL22-22A (CP115858.1). While dcp_2 and cysJ_1 showed 98% and 97% sequence similarities with PL22-22A (CP115858.1), respectively. The multiple-sequence alignments of the keratinolytic enzymes showed variations in specific amino acid residues (Figure 1). Notably, the most variation in the sequence residues was observed from the thioredoxin-dependent 5’-adenylylsulfate reductase amino acid sequence (cysJ_1), where the C. proteolyticum cysJ_1 (MLPAAHBK_00709) showed 90% sequence identity with a similar sequence from Chryseobacterium paludism (WP_261512302.1) and 99% with the unclassified Chryseobacterium (WP_172282549.1) and Chryseobacterium daecheongense (WBV57611.1) (Figure 1a). Variation in amino acid between the recent thioredoxin-dependent 5′-adenylylsulfate reductase amino acid sequence and Chryseo-bacterium paludism amino acid sequence was noticeable in position eleven, where the current sequence had lysine instead of glutamic acid. In position 15, the current sequence exhibited valine, while Chryseobacterium paludism exhibited lysine in this position. Another difference in amino codes in these sequences was observed in position 16, where alanine replaced valine, while in position 19, histidine replaced asparagine, and in position 20, alanine replaced threonine.
Similarly, the multiple sequence alignments of resA (MLPAABK_01466) with other related sequences from Chryseobacterium spp. showed that resA has 98% sequence homology with the amino acid sequences of enzymes from Chryseobacterium sp. B21-037, and 99% sequence similarity with the enzymes from Chryseobacterium sp. CKR4-1 and Chryseobacterium sp. LAM-KRS1 (Figure 1b). The dissimilarity of the amino acid sequences of the resA gene of this study with that of Chryseobacterium sp. B21-037 was observed in two amino acids, including serine in the 45th base, replaced in the query sequence by alanine, and aspartic acid in the 182nd base, replaced by glutamic acid. There was only one dissimilarity noted in the Chryseobacterium sp. LAM-KRS1 and the dissimilarity was at the 19th phenylalanine exchanged by Leucine.
Figure 1. The multiple sequence alignments of the keratinolytic enzymes. (a). Sequence alignments of cysJ_1 (MLPAAHBK_00709) with other highly similar thioredoxin-dependent 5′-adenylylsulfate reductases of the different Chryseobacterium spp. (b). Sequence alignments of resA (MLPAABK_01466) with Chryseobacterium sp. B21-037 (WP_273648577.1), Chryseobacterium sp. LAM-KRS1 (WP_317168537.1), and Chryseobacterium sp. CKR4-1 (WP_273648577.1) enzyme sequences. The variance between sequences is denoted by the colon (:)
Physiochemical properties of the identified keratinolytic genes
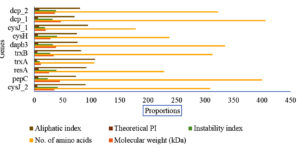
The determination of the physicochemical properties of the keratinolytic enzymes showed that sulfite reductase [NADPH] flavoprotein alpha-component demonstrated a molecular weight of 19.62 kDa, an instability index of 18.83, a theoretical pI of 8.49, aliphatic index of 94.27 (Figure 2). On the other hand, thioredoxin reductase properties were determined to be 33.8 kDa, 27.82, 5.24, and 82.3 for molecular weight, instability index, theoretical pI, and aliphatic index, respectively. Meanwhile, peptidyl carboxypeptidase’s properties computation showed molecular weight, instability index, theoretical pI, and aliphatic index of 46.76 kDa, 31.73, 6.08, and 70.39, respectively (Figure 2). A total of 60% (6/10) of the analysed keratinolytic enzymes (pepC, trxA, trxB, dapb_3, cysH, and dcp_1,) showed to have theoretical pI of more than 7 (Figure 2). On the contrary, 40% (4/10) of the enzymes which include resA, cysJ-1, cysJ_2, and dcp_2 had a theoretical pI of less than 7.
Amplification of selected keratinolytic enzyme-encoding genes from C. proteolyticum genome
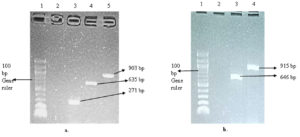
Ten (10) selected genes coding for keratinolytic enzymes were amplified, and the results showed three genes coding for thioredoxin A (trxA), aminopeptidase C (pepC), and dipeptidyl aminopeptidase (dapb_3), with band sizes of 271 bp, 635 bp, and 903 bp, respectively (Figure 3a). Likewise, the amplification of reductases coding genes showed that resA was sitting at 646 bp and trxB was at 915 bp (Figure 3b). The two carboxypeptidases coding genes dcp_1 and dcp_2 were successfully amplified, with a respective band size of 1129 bp and 893 bp (Figure 4a). The genes coding for 5′-adenylylsulfate reductase (cysH) and sulfite reductase (cysJ_1) were also among the amplified genes with band sizes of 562 bp and 409 bp, respectively (Figure 4a). In addition, cysJ_2, which also codes for sulfite reductase, was sitting at 472 bp (Figure 4b).
Figure 3. Gel electrophoresis of the amplified keratinolytic enzyme-encoding genes. (a). Gel picture showing trxA, dapb-3, and pepC genes from C. proteolyticum; Lane 1: 100 bp DNA gene ruler, Lane 2: Negative control, Lane 3: trxA gene (271 bp), Lane 4: dapb-3 (635 bp) gene, and Lane 5: pepC gene (903 bp). (b). Gel picture showing resA and trxB genes; Lane 1: 100 bp DNA gene ruler, Lane 2: Negative control, Lane 3: resA gene (646 bp), and Lane 4: trxB gene (915 bp)
Figure 4. Gel electrophoresis of the amplified keratinolytic enzymes-encoding genes from C. proteolyticum. (a). Gel picture of cysH, cysJ_1, dcp_1, and dcp_1; Lane 1: 100 bp DNA ladder, Lane 2: negative control, Lane 3: cysH (562 bp), Lane 4: cysJ-1 (409 bp), Lane 5: dcp_2 (893 bp), Lane 6: dcp_1 (1129 bp). (b). Gel picture of the cysJ_2 gene; Lane 1: 100 bp DNA ladder, Lane 2: negative control, Lane 3: cysJ_2 (472 bp)
Keratinous waste biomass accumulation is one of the public health challenges seeking urgent attention. The keratinases from different microbial species have proved that these enzyme batteries are effective towards keratinous waste valorisation. Identifying the metabolic pathways involved in the secretion of these enzyme batteries is essential in enhancing their production for large-scale biotechnological products. Hence, this study aimed at mining the C. proteolyticum genome to identify the production of secondary metabolites involved in keratinolysis for prospective biotechnological applications. The revalidation of the keratinolytic potential of the C. proteolyticum strain proves that the complete chicken feather hydrolysis was due to the secretion of keratinolytic proteases. The expressed keratinase activity was similar to the previously reported keratinase activity of characterized C. proteolyticum by Giwu et al,15 who reported 693.63 U/mL. The high keratinase titre demonstrated by C. proteolyticum underscores its robust metabolic diversity and keratinolytic potential.
The presence of free thiols in the media indicates the reduction of disulfide bonds to cysteine thiol and cysteine-S-sulphonate residue, which is facilitated by the series of reductases, including thioredoxin reductases, thioredoxin-dependent 5′-adenylylsulfate reductase, sulfite reductases, and thiol-disulfide oxidoreductases.24-26 The high thiol groups detected in the fermentation broth prove that the chicken feather degradation by C. proteolyticum involved sulfitolysis.17,27,28 In addition, the thiol concentration may indicate the biocatalytic efficiency of the keratinolytic and sulfitolytic systems of keratin-degrading bacteria.17,27,29 The thiol concentration detected in this study was higher than that observed by Kshetri and Ningthoujam29 and He et al.,30 who recorded thiol concentrations of 82 µM and 14.1 µM, respectively, during chicken feather degradation, highlighting that this strain strongly degrades keratin as the carbon and nitrogen source.
The discovery of the keratinolytic enzymes-encoding genes in the genome of C. proteolyticum suggested that keratinolysis is a two-step enzymatic process, which is sulfitolysis and proteolysis, involving enzyme cocktails31 that include reductases, carboxypeptidases, dipeptidases, and aminopeptidases in the bacterial genome.31 During the chicken feather biodegradation, the tightly packed cysteine disulfide bonds are reduced by reductases, leaving the keratin vulnerable to proteolysis by the peptidases.9,10,32,33 The genome profiling further demonstrated that the effective keratin-degrading ability of this bacterium might be associated with the expression of these batteries of genes and cooperative actions of the reductase and peptidase cocktails.34,35 Bacteria genome mining has been identified as an important approach that has been effectively employed in discovering important secondary metabolites with keratinolytic dexterity.5
Multiple-sequence alignment helps to identify the evolutionary relationships among sequences with similar biological activity.36 The disparity in amino acid residues at some positions on the aligned sequences might be attributed to the bacterial strain variations, and this residue substitution could suggest the uniqueness of the keratinolytic enzymes under investigation.17 The observed sequence divergence could also influence the catalytic tendency of the biocatalysts towards substrate utilization. In silico determination of bacterial enzyme’s physicochemical properties provides interesting data relevant to the enzyme application potential. The aliphatic index is one of the essential physicochemical properties that suggest a protein’s thermal stability profile, and a protein with a high aliphatic index is more thermally stable than those with a low aliphatic index.37 All the investigated keratinolytic enzymes displayed prospects of thermal stability, with an aliphatic index between 70 and 106. Additionally, the possessed aliphatic index proves that these enzymes consisted of hydrophobic amino acids such as alanine, isoleucine, leucine, aspartic acid, and valine.38 The observed theoretical pI was between 4 units, suggesting that these enzymes would show activity optima from acidic to a basic region of the pH spectrum. According to Halligan,39 a theoretical pI value greater than 7 suggests that the protein or enzyme is acidic.
The successful amplification of the selected keratinolytic enzymes-encoding genes suggests the feasibility of manipulating the isolate’s keratinolytic determinants and their subsequent use in the transformation of industrially competent cells through genetic engineering to enhance keratinolytic activity. Scalable production of keratinolytic enzymes will not only promote sustainable valorization of the recalcitrant agroindustrial waste but also revolutionize the application of the enzymes in different sectors of the bioeconomy.40
The genome of a competent feather-degrading bacterium, C. proteolyticum, was sequenced and annotated to identify its keratinolytic determinants. Out of the 4798 genes obtained through Prokka annotation, thirty-eight proved to be putative keratinolytic enzyme-encoding genes. Multiple-sequence alignment demonstrated that the amino acid sequences of the putative keratinolytic enzymes displayed some variations with related sequences retrieved from the protein database. In silico analysis of the physicochemical characteristics showed that the enzymes displayed variable properties with a high thermostability profile. Successful amplification of the selected putative keratinolytic enzyme-encoding genes from the C. proteolyticum genome highlights its endowed keratinolytic tendencies for the sustainable valorization of keratinous biomass. The results of this study confirm the feasibility of heterologous expression of the identified genes in competent hosts to upscale the keratinolytic enzyme production and demand further investigation.
ACKNOWLEDGMENTS
The authors would also like to appreciate the support from the Infectious Diseases and Medicinal Plants Research Niche Area, University of Fort Hare, Alice, South Africa.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
LM, NEN and UUN conceptualized the study. UUN performed funding acquisition. UUN collected resources. LM applied methodology. LM and NEN performed formal analysis and investigation. LM wrote the original draft. NEN and UUN wrote, reviewed and edited the manuscript. NEN and UUN performed supervision. All authors read and approved the final version of the manuscript for publication.
FUNDING
This work was supported by the Industrial Biocatalysis Hub, Department of Science and Innovation, and the Technology Innovation Agency, South Africa.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Li Q. Progress in microbial degradation of feather waste. Front Microbiol. 2019;10:2717.
Crossref - Wang L, Zhou Y, Huang Y, Wei Q, Huang H, Guo C. Cloning and expression of a thermostable keratinase gene from Thermoactinomyces sp. YT06 in Escherichia coli and characterization of purified recombinant enzymes. World J Microbiol Biotechnol. 2019;35(9):135.
Crossref - Cavello IA, Cavalitto SF, Hours RA. Biodegradation of a keratin waste and the concomitant production of detergent stable serine proteases from Paecilomyces lilacinus. Appl Biochem Biotechnol. 2012;167(5):945-958.
Crossref - Wu WL, Chen MY, Tu IF, et al. The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles. Sci Rep. 2017;7(1):4658.
Crossref - Fellahi S, Chibani A, Feuk-Lagerstedt E, Taherzadeh MJ. Identification of two new keratinolytic proteases from a Bacillus pumilus strain using protein analysis and gene sequencing. AMB Expr. 2016;6(1):42.
Crossref - Kalaikumari SS, Vennila T, Monika V, Chandraraj K, Gunasekaran P, Rajendhran J. Bioutilization of poultry feather for keratinase production and its application in leather industry. J Clea Prod. 2019;208:44-53.
Crossref - Nnolim NE, Nwodo UU. Microbial keratinase and the bio-economy:a three-decade meta-analysis of research exploit. AMB Expr. 2021;11(1):1-16.
Crossref - Fagbemi OD, Sithole B, Tesfaye T. Optimization of keratin protein extraction from waste chicken feathers using hybrid pre-treatment techniques. Sustain Chem Pharm. 2020;17:100267.
Crossref - Lange L, Huang Y, Busk PK. Microbial decomposition of keratin in nature-A new hypothesis of industrial relevance. Appl Microbiol Biotechnol. 2016;100(5):2083-2096.
Crossref - Peng Z, Zhang J, Du G, Chen J. Keratin waste recycling based on microbial degradation: mechanisms and prospects. ACS Sustain Chem Eng. 2019;7(11):9727-9736.
Crossref - Erlendsson LS, Moller M, Hederstedt L. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J Bacteriol. 2004;186(18):6230-6238.
Crossref - Kouwen TRHM, Dubois JF, Freudl R, Quax WJ, van Dijl JM. Modulation of thiol-disulfide oxidoreductases for increased production of disulfide-bond containing proteins in Bacillus subtilis. Appl Environ Microbiol. 2008;74(24):7536-7545.
Crossref - Gonzales T, Robert-Baudouy J, Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev. 1996;18(4):319-344.
Crossref - Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Med Mycol. 2019;57(1):13-22.
Crossref - Giwu N, Nnolim NE, Nwodo UU. Keratinases produced by Chryseobacterium proteolyticum FGNn and Pseudomonas aeruginosa GNFx liberated amino acids from poultry feathers. Biomass Convers Biorefin. 2023;1-1315(1):687-699.
Crossref - Letourneau, Soussotte, Bressollier, Branland, Verneuil, Keratinolytic activity of Streptomyces sp. S. K1-02: a new isolated strain. Lett Appl Microbiol. 1998;26(1):77-80.
Crossref - Nnolim NE, Nwodo UU. Bacillus sp. CSK2 produced thermostable alkaline keratinase using agro-wastes:keratinolytic enzyme characterization. BMC Biotechnol. 2020;20(1):1-14.
Crossref - Jaouadi B, Abdelmalek B, Fodil D, et al. Purification and characterisation of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour Technol. 2010;101(21):8361-8369.
Crossref - Sunnucks P, Hales DF. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). MBE. 1996;13(3):510-524.
Crossref - Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinform. 2020;70(1):e102.
Crossref - Mikheenko A, Valin G, Prjibelski A, Saveliev V, Gurevich A. Icarus: visualizer for de novo assembly evaluation. Bioinform. 2016;32(21):3321-3323.
Crossref - Seemann T. Prokka:Rapid prokaryotic genome annotation. Bioinform. 2014:30(14):2068-2069.
Crossref - Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker, J.M. (eds) The Proteomics Protocols Handbook. Totowa, NJ: Humana Press. 2005;571-607.
Crossref - Ramnani P, Singh R, Gupta R. Keratinolytic potential of Bacillus licheniformis RG1 is structural and biochemical mechanism of feather degradation. Can J Microbiol. 2005:51(3):191-196.
Crossref - Ghosh A, Chakrabart K, Chattopadhyay D. Degradation of raw feather by a novel high molecular weight extracellular protease from newly isolated Bacillus cereus DCUW. J Ind Microbiol Biotechnol. 2008;35(8):825-834.
Crossref - Kumar AG, Swarnalatha S, Gayathri S, Nagesh N, Sekaran G. Characterization of an alkaline active-thiol forming extracellular serine keratinase by the newly isolated Bacillus pumilus. J Appl Microbiol. 2008;104(2):411-419.
Crossref - Brandelli A, Daroit DJ, Riffel A. Biochemical features of microbial keratinases and their production and applications. Appl Microbiol Biotechnol. 2010;85(6):1735-1750.
Crossref - Kshetri P, Roy SS, Sharma SK, et al. Transforming chicken feather waste into feather protein hydrolysate using a newly isolated multifaceted keratinolytic bacterium Chryseobacterium sediminis RCM-SSR-7. Waste Biomass Valori. 2019;10(1):1-11.
Crossref - Kshetri P, Ningthoujam DS, Keratinolytic activities of alkaliphilic Bacillus sp. MBRL 575 from a novel habitat, limestone deposit site in Manipur, India. SpringerPlus. 2016;5:1-16.
Crossref - He Z, Sun R, Tang Z, et al. Biodegradation of feather waste keratin by the keratin-degrading strain Bacillus subtilis 8. 2018;28(2):314-322.
Crossref - Yamamura S, Morita Y, Hasan Q, et al. Characterization of a new keratin-degrading bacterium isolated from deer fur. J Biosci Bioeng. 2002;93(6):595-600.
Crossref - Sharma R, Rajak RC. Keratinophilic Fungi:Nature’s keratin degrading machines! Their isolation, identification and ecological role. Reson. 2003;8(9):28-40.
Crossref - Singh I, Kushwaha RKS. Keratinases and microbial degradation of keratin. Adv Appl Sci Res. 2015;6(2):74-82.
- Kang E, Jin HS, La JW, et al. Identification of keratinases from Fervidobacterium islandicum AW 1 using dynamic gene expression profiling. Microb Biotechnol. 2020;13(2):442-457.
Crossref - Ji X, Yang J, Chen J, Zhang J, Peng Z. A Synergistic catalytic system of keratinase and disulfide reductase for the bioconversion of feather wastes to feed alternative protein. Food Biosci. 2024;59:104219.
Crossref - Jia K, Kilinc M, Jernigan RL. New alignment method for remote protein sequences by the direct use of pairwise sequence correlations and substitutions. Front Bioinform. 2023;3:1227193.
Crossref - Panda S, Chandra G. Physicochemical characterization and functional analysis of some snake venom toxin proteins and related non-toxin proteins of other chordates. Bioinformation. 2012;8(18):891-896.
Crossref - Pal S, Sengupta K. Computational-based insights into the phylogeny, structure, and function of Rhodococcus alkane-1-monooxygenase. 3 Biotech. 2020;10(9):391.
Crossref - Halligan BD. ProMoST:a tool for calculating the PI and molecular mass of phosphorylated and modified proteins on two-dimensional gels. Phospho-Proteomics:Methods and Protocols. 2009;527:283-298.
Crossref - Rahimnahal S, Meimandipour A, Fayazi J, et al. Biochemical and molecular characterization of novel keratinolytic protease from Bacillus licheniformis (KRLr1). Front microbiol. 2023;14:1132760.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.