ISSN: 0973-7510
E-ISSN: 2581-690X
Millions of people have died due to coronavirus infection, which has also brought an unparalleled global health crisis to the verge of collapsing several nations healthcare system. The host genetic landscape is becoming more widely acknowledged as a crucial determinant of susceptibility to infection among all other factors. It is still unclear how exactly they are related, though. As a result, we looked into the significance of genetic factors in COVID-19 patients’ severity by performing a thorough analysis of research that were documented in the literature. A thorough search of PubMed was started at the beginning of July 2021. From December 2019 to July 2021, we retrieved all pertinent papers and screened on inclusion and exclusion criteria. This systematic review comprised twenty-five papers with COVID-19 case studies for qualitative synthesis. Seven different gene types, including human leukocyte antigen (HLA), angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), ABO, Cluster of differentiation 45 (CD45), open reading frame (ORF) and interleukin-6 (IL-6) genes were identified, which might be responsible for the significance of coronavirus illness in the patients. Overall, individuals with COVID-19 who also possess the aforementioned genes are at a higher risk of developing significant illness. Still, more information is needed to draw a reliable conclusion.
SARS-CoV-2, Genetics, ACE, IL-6, Gene, Mortality, Disease, Illness
SARS-CoV-2 viral infection created a havoc situation across the globe and presented hitherto unheard-of difficulties for global healthcare and economic systems. This was initially found in Wuhan, Hubei Province, China, in December 2019.1,2 The World Health Organising (WHO) has classified the disease as both a pandemic and public health emergency, implying a global mortality rate of 2.22 percent.1-3 Depending on the availability of diagnostic technologies, several countries adopted and implemented different testing procedures during the early days of the epidemic.3 The degree of symptoms varies greatly among people, ranging from asymptomatic to a serious condition with deadly implications. One of the main questions that confounded healthcare professionals and researchers during the COVID-19 pandemic’s unfolding was why some people experienced severe, frequently fatal consequences while others only experienced minor disease or showed no symptoms.4 Some researchers believe that different immune cells in the body can help with infection.4 On the other hand, the remarkable scientific research to COVID-19 has elucidated the disease pathogenesis and pinpointed the risk factors that contribute disproportionately to the progression of severe illness. All age groups are susceptible to catching SARS-CoV-2, but older people and those with comorbid conditions such as diabetes, hypertension, obesity, cardiovascular disease, lung and kidney disease, cancer, organ transplant, and weak immune system are more likely to experience symptoms.1,5 Furthermore, a sudden increase in pro-inflammatory cytokines may be lethal, indicating a potential involvement for cytokines in COVID-19 pathogenesis.5-7 Thus, it is vital to study the effect of genetic polymorphism on the pathogenesis of COVID-19. The main aim of the current study is to find out the human genetic variations contributing COVID-19 Intensity. In terms of improving our knowledge of the host genetic foundation of COVID-19 symptoms, we anticipate that the results of this study will help identify possible therapeutic targets and help develop personalised treatment plans.
Search strategy
For a literature search with particular MeSH standings, electronic databases including PubMed, Google Scholar, and SciHub were explored. The following terms were used: “Gene” OR “SNP” OR “Single nucleotide polymorphism OR Genetic variability” OR “Genome” OR “Exome” OR “Sequencing” AND “NRPI” OR “TFTM3” OR “LZTFL” OR “ABO” OR” IL-6″ OR “IFNY” AND “COVID” OR “Corona” OR “NOVEL CORONA VIRUS” OR “SARS-COVID-19” OR “SARS-CoV-2” OR “COVID-19” OR “SARS associated coronavirus 2” OR “severe acute respiratory syndrome-2”. The studies were included from 1st December 2019 to 14th July 2021. Further, the Preferred Reporting Items for Systematic Reviews and Meta-analysis Standards (PRISMA) was used for data extraction.8
Eligibility criteria
The following detailed eligibility criteria followed for the extraction of studies from the literature.
Inclusion criteria
- All study types, multicentre studies, controlled clinical trials, randomised controlled trails, comparative studies, and pragmatic clinical trials are among the inclusion criteria.
- Articles outlining the clinical traits of coronavirus patients.
- Articles outlining COVID-19 severity and contributing variables.
Exclusion criteria
- Publications published in languages other than English are excluded.
- Editorials, case reports, review articles, meta-analyses, letters, and comments or opinions.
- Articles with incomplete information.
Study selection
Two authors (MI and A) independently screen the literature to minimize the bias. Following that, the entire texts of selected publications were thoroughly evaluated using the inclusion and exclusion criteria. Third Author (RMAA) was consulted about any disagreement over the inclusion of data extraction.
Data mining
Two different authors extracted the data using consistent and standard forms, which included authors, place, type of study, date of publication, total sample size, sex, and severe findings.
Quality assessment
NOS was used by two authors (MI, A) to check the quality of included studies. A 7 out of 10 on the quality grading scale denotes exceptional excellence. For the statistics and the scale from 0 to 10, 0-3 was terrible, 3-5 was fair, and 5-7 was good. Third Author (RMAA) was consulted about any disagreement.
Search Strategies and Study Preference
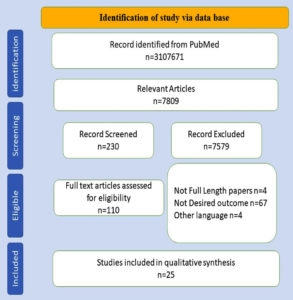
Duplicates were discarded after an initial evaluation of the retrieved resources; further, on the basis of the title and abstract, the articles were screened. The initial search techniques returned 31,07,671 items, of which 7809 were included after the eligibility requirements were assessed using filters. Out of the 230 manuscripts that were reviewed based on the abstract, only 110 were deemed suitable for data extraction. Finally, 25 publications were considered that were published between 1st December 2019 and July 14th, 2021, and were clinical and retrospective cohort studies.9-33 The sorting of studies is displayed in Figure.
Quality evaluation
The 25 articles were included for the quality assessment using the Newcastle-Ottawa scale. All of the 25 articles were of excellent quality as described in Table 1.
Table (1):
Quality assessment
Reference |
Selection |
Compatibility |
Outcome |
Total score |
Quality of the study |
|---|---|---|---|---|---|
Asselta et al.9 |
**** |
** |
** |
8 |
Excellent |
Benetti et al.10 |
**** |
** |
*** |
9 |
Excellent |
Camiolo et al.11 |
**** |
* |
**** |
9 |
Excellent |
Mao et al.12 |
**** |
* |
*** |
7 |
Excellent |
Palazzuoli et al.13 |
**** |
** |
*** |
9 |
Excellent |
Tang et al.14 |
**** |
** |
**** |
8 |
Excellent |
Srivastava et al.15 |
**** |
* |
*** |
8 |
Excellent |
Wallentin et al.16 |
**** |
** |
**** |
10 |
Excellent |
Azevedo et al.17 |
**** |
** |
*** |
9 |
Excellent |
D’alessandro et al.18 |
**** |
* |
**** |
9 |
Excellent |
Della-Torre et al.19 |
**** |
** |
*** |
9 |
Excellent |
Galvan-Roman et al.20 |
**** |
** |
*** |
9 |
Excellent |
Sadeghi et al.21 |
**** |
** |
*** |
9 |
Excellent |
Vultaggio et al.22 |
**** |
** |
*** |
9 |
Excellent |
Wang et al.23 |
**** |
** |
*** |
9 |
Excellent |
Xia et al.24 |
**** |
** |
*** |
9 |
Excellent |
Yuan et al.25 |
**** |
** |
*** |
9 |
Excellent |
Hoiland et al.26 |
**** |
** |
*** |
9 |
Excellent |
Niles et al.27 |
**** |
** |
*** |
9 |
Excellent |
Naufal et al.28 |
**** |
* |
*** |
8 |
Excellent |
Sardu et al.29 |
**** |
** |
*** |
9 |
Excellent |
Correale et al.30 |
**** |
* |
**** |
9 |
Excellent |
Lorente et al.31 |
**** |
** |
*** |
9 |
Excellent |
Young et al.32 |
**** |
* |
*** |
8 |
Excellent |
Jin et al.33 |
**** |
** |
*** |
9 |
Excellent |
Association of genes with the seriousness of COVID-19
The genetic makeup of an individual determines their susceptibility to viral infection and how they respond to it. However, the severity of COVID-19 disease is also a result of other factors, including social, environmental, and clinical ones.34-36 In order to further genetic discovery for SARS-CoV-2 infection and severe sickness, experts from all around the world have joined forces for the COVID-19 host genetics study.37-39 Seven gene categories were found to affect the severity of confirmed cases. The detailed explanation is given below.
Angiotensin-converting enzyme 2 (ACE2)
The ACE2 gene has 18 exons and is situated on chromosome Xp22.40 ACE2 serves a variety of functions, including catalytic activity with specific substrates, negative regulation of the RAAS, and coronavirus receptor. COVID-19 may have a pathogenic mechanism akin to SARS because both SARS-CoV-1 and 2 spoke protein interacts with ACE2.38-41
There are eight published research that look at the association of severity of infection with ACE2. Wallentin et al. conducted two large cohort studies to look at the connection between COVID-19 patients’ severity and the ACE2 gene. The results of this investigation show a robust correlation between ACE2 and the likelihood of death and severe COVID-19 infection.16 However, the Palazzuoli et al. study indicated that renin-angiotensin-aldosterone system inhibition protects COVIS-19 patients and reduce the severity of COVID-19 in patients with high cardiovascular risk.13 Furthermore, Benetti et al., did a cohort study in the Italian population to show a strong correlation between ACE2 gene variant and interindividual variability for COVID-19 susceptibility. Their findings suggest that a predisposed genetic background could play a role in observing COVID-19 interindividual clinical heterogeneity.10 However, Asselta et al., used ACE2 expression levels and variations to undertake a cohort analysis to uncover probable genetic components for COVID-19 severity in Italians. Despite this, their research did not identify a significant link between illness severity and ACE2 expression.9 Table 2 summarises the research that links ACE2 expression to COVID-19 patient severity.
Table (2):
Role of ACE2 during COVID-19
No. |
Author |
Place |
Study type |
Male |
Female |
Total no. of subjects |
Findings |
|---|---|---|---|---|---|---|---|
1 |
Asselta et al.9 |
Italy |
Cohort study |
395 |
183 |
578 |
There is no evidence that ACE2 is linked to illness severity. However, genetic factors influence COVID-19 severity, with a focus on ACE2-mediated effects. |
2 |
Benetti et al.10 |
Italy |
Cohort Study |
97 |
34 |
131 |
According to the results, there could be a genetic predisposition contributing to the interindividual clinical heterogeneity linked to COVID-19. |
3 |
Camiolo et al.11 |
USA |
Cohort study |
– |
– |
– |
In asthma patients with characteristics that resemble established risk factors severe infection and the expression of ACE2 is associated with viral gene. |
4 |
Mao et al.12 |
USA |
Clinical study |
– |
– |
14 |
Pathogenesis and the increase of ACE2 expression are linked to placental hypoxia. |
5 |
Palazzuoli et al.13 |
Italy |
Clinical study |
498 |
– |
781 |
Association of ACE2 with the severity of COVID-19 patients. Also, reported the protective role of suppression of the RAAS pathways. |
6 |
Tang et al.14 |
Russia |
Clinical study |
552 |
259 |
811 |
Significant symptoms may be more likely to occur in those with KIRC or KIRP due to the ACE2 expression in kidney carcinoma and SARS-Cov-2 infection. |
7 |
Srivastava et al.15 |
India |
Observational study |
– |
– |
248 |
They found that the reference allele has a 50 percent lower ACE2 expression, resulting in severity of the infection |
8 |
Wallentin et al.16 |
USA, Canada, Sweden, Germany, France |
Cohort study |
3202 |
– |
5087 |
The biomarkers GDF-15 and BNP were most strongly related to the level of ACE2. An increase in these indicators raises the likelihood of death and severe COVID-19 infection. |
Interleukin-6 (IL-6)
Determining the nature and classification of the cytokine storm is a difficult task. On the other hand, it describes an uncontrolled inflammatory response and an overactive immune system, both of which have a wide range of negative health effects and are linked to both infectious and non-infectious illnesses.42 Numerous immune cells become activated in response to the SARS-CoV-2 infection, releasing a variety of cytokines, including the pro-inflammatory cytokine IL-6.43 Along with the hormone-like characteristics of vascular illness, this cytokine also impacts mitochondrial activity, insulin resistance, lipid metabolism, the neuroendocrine system, and cognitive behaviour. It accomplishes this by encouraging B-cell differentiation and T-cell proliferation. Additionally, vascular endothelial cells and the coagulation cascade can be activated by massive doses of IL-6.44
There are nine research that we could locate that have attempted to determine how important the IL-6 gene is for COVID-19 patients’ severity. A clinical investigation by Wang et al. was carried out in China to determine the relationship between inflammatory and non-inflammatory markers with the severity of the infection. Researchers found that IL-6 levels were greater in severe COVID-19 patients, and they suggest that IL-6 is a good proxy for severity in COVID-19 patients. Important components of the adaptive immune system, Th17 and Treg cells release IL-6 among other cytokines. In 2020, Sadeghi et al. did a clinical experiment to examine Th17 and Treg cell responses in COVID-19 patients, and reported direct relationship of IL-6 with a severity of COVID-19 patients. The results of the Galvan-Roman et al. investigation also revealed a relationship between the IL-6 level and the severity of COVID-19 patients’ condition. A robust correlation has been observed between IL-6 and the elevated mortality risk in COVID-19 patients.20 Results from the Chinese Yuan group in 2021 likewise showed a strong relationship between the severity of COVID-19 and cytokine production.25 Table 3 presents the information about IL-6 and the severity of patients.
Table (3):
Role of IL-6 during COVID-19
No. |
Author |
Place |
Type of Study |
Male |
Female |
Total sample size |
Findings |
|---|---|---|---|---|---|---|---|
1 |
Azevedo et al17 |
Brazil |
Clinical study |
13 |
7 |
20 |
No stastical differences were observed regarding IL-17A gene’s genotype frequencies. In COVID-19 patients who passed away, the G allele in SNP rs3819025 (G/A) may be regarded as a risk allele. |
2 |
D’alessandro et al.18 |
Columbia |
Clinical study |
– |
– |
33 |
inhibitory components of the fibrinolytic cascade were found to be increased in severe infection. |
3 |
Della-Torre et al.19 |
Italy |
Cohort study |
124 |
– |
210 |
They found if drug administered before to the onset of severe respiratory failure, IL-1 and IL-6 blocking medications had a greater chance of improving survival in hyper-inflamed COVID-19 patients. |
4 |
Galvan-Roman et al.20 |
Spain |
Retrospective observational study |
97 |
– |
146 |
Since tocilizumab (TCZ) blocks the receptor for IL-6, it may be possible to reduce mortality and/or morbidity in COVID-19 patients with severe illness. |
5 |
Sadeghi et al.21 |
Iran |
Clinical study |
28 |
12 |
40 |
In COVID-19 patients, aberrant Th17/Treg cell ratios, Treg cells, and their related factors may be important in promoting inflammatory responses and the development of the illness. |
6 |
Vultaggio et al.22 |
Italy |
Retrospective observational study |
134 |
74 |
208 |
As the quartiles of IL-6 levels increased, the likelihood of clinical deterioration increased as well. |
7 |
Wang et al.23 |
China |
Clinical study |
49 |
28 |
77 |
The levels of blood cTnI, D-D, CRP, IL-6, PCT, neutrophil, and lymphocyte counts were significantly different between the severe and critical COVID-19 patients. |
8 |
Xia et al.24 |
China |
Retrospective, observational study |
161 |
– |
330 |
High-sensitivity troponin I was discovered to have a significant positive correlation with inflammatory cytokines. Elevations in inflammatory cytokines have been associated with heart injury in COVID-19 patients who are severely and critically unwell. |
9 |
Yuan et al.25 |
China |
Retrospective study |
63 |
56 |
119 |
The findings show that there is a strong link between therapy can lessen lung damage in patients by inhibiting excessive cytokine production. |
Transmembrane serine protease 2 (TMPRSS2)
The pathophysiology of SARS-CoV and SARS-CoV-2 appears to include the TMPRSS2. Furthermore, TMPRSS2 has been detected in the aerodigestive tract, even though the prostate epithelium expresses it multiple times more than any other tissue.17 Interestingly, variations in TMPRSS2 expression in lung cells may exist between groups, suggesting a role for influenza and coronavirus infection susceptibility. Determining how the TMPRSS2 protein is expressed in the lungs is therefore crucial to comprehending the SARS-CoV-2 infection susceptibility between men and women.9 Nonetheless, conflicting findings have been obtained by other studies, most of which maintain that gender has no bearing on the constitutive expression of TMPRSS2 in lung cells.17 The risk, severity, and clinical consequences of COVID-19 have been linked to genetic polymorphisms in the ACE2 and TMPRSS2 genes. Only one study has looked at the association between TMPRSS2 and the severity of COVID-19 patients. Asselta et al. used cohort research to investigate expression levels and variations in TMPRSS2 to uncover putative genetic components for COVID-19 severity in Italians. In their cohort research, they included 578 patients (395 men and 183 women). Despite this, their research found no significant link between disease severity and TMPRSS2 expression. However, TMPRSS2 levels and genetic variations, on the other hand, were discovered to represent potential disease modulators.9
ABO
Research on ABO has demonstrated its significance in COVID-19 susceptibility and clinical symptoms, both in hereditary and non-genetic contexts.45 According to earlier research and genome-wide association studies (GWAS), people in the O blood type are less likely to become infected with COVID-19, whereas people in the A blood group are more likely to do so than people in other blood groups.46 Previous studies have linked the ABO blood group to a higher chance of catching influenza, schistosomiasis, malaria, SARS-CoV-2, and malaria. According to the theory, blood antigens can serve as either receptors or co-receptors for microorganisms. They can also facilitate intracellular absorption, adhesion, and signal transmission.47,48 Moreover, it has been proposed that the innate immune response to the virus may involve naturally occurring antibodies linked to blood types. However, it is still unclear what exact function the ABO groups play in the SARS-CoV-2 infection pathway.
Niles et al. conducted female cohort research in Columbia to examine the association between COVID-19 positive and ABO/Rh. Relationships between COVID-19 positive and severity, ABO group, and Rh types were found in their investigation.27 Similarly, Hoiland et al., did a clinical investigation in Columbia to see if there was a link between severity and blood group. Their findings demonstrated a substantial link between COVID-19 patients with blood groups A or AB and those who were seriously ill.26 Furthermore, Naufal et al. investigated the association between ABO blood type and COVID-19 mortality through cohort research carried out in the United States. Their research showed a correlation between the blood types of COVID-19 patients and their risk of cardiovascular issues.28 Table 4 summarises studies associated with blood groups in relation to the seriousness of COVID-19 patients.
Table (4):
Role of ABO during COVID-19
No |
Author |
Place |
Type of study |
Male |
Female |
Total sample size |
Findings |
|---|---|---|---|---|---|---|---|
1 |
Hoiland et a.l26 |
Columbia |
Clinical Study |
61 |
34 |
95 |
Findings show that critically ill COVID-19 patients with blood groups A or AB are more likely a longer stay in the ICU. |
2 |
Niles et al.27 |
Columbia |
Cohort study |
– |
88975 |
88975 |
The study looked into the links between ABO group/Rh types and SARS-CoV-2 positive among people of different races and ethnicities. |
3 |
Naufal et al28 |
USA |
Cohort study |
– |
– |
1114 |
In patients with COVID-19, blood group A was linked to an increased risk, whereas blood group O was linked to a lower risk. These findings could help to stratify COVID-19 patients’ risk of cardiovascular problems. |
4 |
Sardu et al.29 |
Italy |
Clinical study |
108 |
– |
164 |
In hypertensive patients with COVID-19 infection, the ABO blood type is linked to a worse outcome. As a result, we believe that in order to prevent heart injury and death in these high-risk COVID-19 patients, specifically hypertensive adults with non-O blood groups, targeted anticoagulant treatments should be introduced early. |
Human leukocyte antigen (HLA)
Numerous HLA alleles have been linked to the severity and susceptibility of COVID-19 by various techniques and in subpopulations.49 The disease-associated HLA allele may occasionally be common across several groups. For example, the HLA-A*11 gene was found in a sample of Chinese and Spanish research participants, but not with Italian patients. Consequently, when looking for the COVID-19 genetic marker, interethnic diversity in HLA allele frequencies should be taken into account.50,51 Moreover, since the viral mutation sites alter how well HLA molecules show antigen, it is imperative to look into how SARS-CoV-2 genome variants affect relevant host alleles. An observational and prospective study by Lorente et al. examined the association between HLA genetic variations and the mortality and vulnerability of COVID-19 patients. HLA genetic variants were shown to be linked to COVID-19 mortality in their study.31 A cohort research was conducted in the Italian population by Correale et al. to ascertain the association between COVID-19 and HLA. According to their findings, pathogenesis is significantly influenced by HLA genes.30 Table 5 presents a comprehensive examination of the relationship between the HLA gene and the severity of COVID-19 patients.
Table (5):
Role of HLA during COVID-19
No. |
Author |
Place |
Type of Study |
Male |
Female |
Total sample |
Findings |
|---|---|---|---|---|---|---|---|
1 |
Correale et al.30 |
Italy |
Cohort study |
– |
– |
370,000 |
HLA-C01 and B*44 alleles play a positive role in COVID-19 pathogenesis. |
2 |
Lorente et al.31 |
Spain |
Retrospective observational study |
38 |
34 |
72 |
COVID-19 mortality may be linked to HLA genetic variations. |
Open reading frame (ORF)
The Open reading frame (ORF) 1a/1ab polyprotein and four structural proteins expressed by CoV genomes have been studied as possible therapeutic targets.52 Additionally, the genomes have varying numbers of ORF encodes those proteins that appear to be important in pathogenesis but are not required for virus replication. Conventional bioinformatics methods have a tougher time predicting accessory proteins since they are less well understood.53 In a cohort experiment conducted in Singapore, Young et al. looked into how the severity of the infection and the inflammatory response were affected by a significant loss in the SARS-CoV-2 genome. Based on their cohort analysis, which included 131 patients in total, they concluded that ORF8 is the hotspot for the severity of COVID-19.33
Cluster of differentiation 45 (CD45)
The common antigen of lymphocytes is the PTPRC, which is unique to leukocytes. It affects TCR activation in two contradictory ways.54 Tyrosine 394 (Y394) is dephosphorylated by CD45, which inhibits the activation of lymphocyte-specific protein tyrosine kinase. On the other hand, when tyrosine 505, an inhibitory location at the C-terminal end of non-receptor tyrosine-Src kinases, is dephosphorylated, CD45 acts as an activator. The immunoreceptor tyrosine-based activation motifs of the TCR/CD3 complex are phosphorylated by activated lymphocyte-specific protein tyrosine kinase.55 Jin et al. conducted a clinical investigation on 331 COVID-19 patients, comprising 175 males and 156 females, in order to determine the role of CD45 in the severity of COVID-19 patients. The findings of the study indicated that CD45 has an effect on the severity of COVID-19 patients, and that immune system malfunction is a likely cause of mortality.33
We searched extensively for this review, encompassing all studies that examined the connection between genetic variations and the likelihood of acquiring SARS-CoV-2 and experiencing severe COVID-19. During the screening process, 7809 articles in total were included, and 25 publications were ultimately taken into consideration for this systematic review. The result revealed that polymorphism between 7 gene, including human leukocyte antigen (HLA), angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), ABO, cluster of differentiation 45 (CD45), open reading frame (ORF) and interleukin-6 (IL-6) genes, it may be accountable for the COVID-19 patients’ severity. These investigations, however, only covered a portion of the possible genetic indicators that have been evaluated for their ability to increase COVID-19 risk. The quantity of genetic association studies on COVID-19 has increased dramatically in recent years, necessitating the compilation of all published data. However, social, environmental, and clinical factors can contribute to the severity of COVID-19 disease.34,56
TMPRSS2 and ACE2 serve as receptor and S protein priming factors, respectively, in the cellular entrance of SARS-CoV-2.39 In addition to being a receptor for the SARS CoV-2 entryway, ACE2 also regulates the ACE1-mediated renin-angiotensin system (RAS). As such, alterations to the ACE1/ACE2 expression levels or signalling axis may be the cause of COVID-19 symptoms.38 Confounders in COVID-19 spread were identified as polymorphisms in TMPRSS2, and ACE.41,42 Correspondingly, Wallentin et al. conducted two large cohort studies to look at the connection between COVID-19 patients’ severity and the ACE2 gene. The results of this investigation show a robust correlation between ACE2 and the likelihood of death and severe COVID-19 infection.16 However, the Palazzuoli et al. study indicated that renin-angiotensin-aldosterone system inhibition protects COVID-19 patients and reduce the severity in patients with high cardiovascular risk.13 Interestingly, variations in TMPRSS2 expression in lung cells may exist between groups, suggesting a role for influenza and coronavirus infections susceptibility. Determining how the TMPRSS2 protein is expressed in the lungs is therefore crucial to comprehending the variations in COVID-19 susceptibility between men and women.9
It has been discovered that cytokine storms mostly contribute to the severity of COVID-19 cases. Numerous immune cells are activated by the SARS-CoV-2 infection, generating a variety of cytokines, including the pro-inflammatory cytokine IL-6.43 This cytokine affects mitochondrial activity, insulin resistance, lipid metabolism, the neuroendocrine system, and cognitive behaviour in addition to the hormone-like features of vascular disease. Additionally, vascular endothelial cells and the coagulation cascade can be activated by high concentrations of IL-6.44 There are nine research that we could locate that have attempted to determine how important the IL-6 gene is for COVID-19 patients’ severity. Sadeghi and coworkers did a clinical experiment to examine Th17 and Treg cell responses in SARS-CoV-2 patients, and found increased level of IL-6 was related with a higher severity infection.
Previous research has linked the ABO blood group to a higher chance of contracting influenza, schistosomiasis, malaria, and SARS-CoV-2 infections. The hypothesis states that blood antigens can function as co-receptors or receptors for microorganisms. In 2020, Niles et al. carried out female cohort studies in Columbia to investigate the relationship between ABO/Rh and COVID-19 positivity. Their analysis revealed relationships between ABO group, Rh types, and COVID-19 positivity and severity.27
Polymorphism in HLA allele frequencies observed while searching for the COVID-19 genetic marker.50,51 An observational and prospective study by Lorente et al. examined the relation of HLA polymorphism with COVID severity and the mortality of SARS-CoV-2 patients.31 Based on their cohort study, Young and coworkers observed that ORF8 is one of the hotspots for the severity of COVID-19.33 Jin et al. carried out a clinical examination in a Chinese study to ascertain the role of CD45 in the severity of COVID-19 patients. They discovered that immune system failure is most likely the cause of death in COVID-19 patients and that CD45 influences the severity of the virus.33
However, the findings of this systematic review advance our knowledge of the host genetic factors that influence the severity and susceptibility of COVID-19. Even though the literature was thoroughly searched, it is possible that some relevant papers were missed. To locate the most recent reports, we manually reviewed the published papers’ reference lists in-depth and repeatedly updated the search, reducing the chance that any relevant articles would be overlooked. Studies conducted in other languages and unpublished data were excluded from our search because they might have influenced the results because we restricted it to published articles written in English.
Seven genes are known to possess a major influence in the severity of COVID-19, according to available data. Further epidemiological research with a larger sample size in genetically diverse groups is still necessary to validate our findings. To understand the fundamental biological pathways, identify the vulnerable individuals, and open the door to developing effective COVID-19 therapy options.
ACKNOWLEDGMENTS
The authors would like to thank INTI International University, Negeri Sembilan, Malaysia, for funding their study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by INTI International University, Malaysia.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- WHO (World Health Organization). WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ (Accessed on Dec 17, 2022).
- Siordia JAJr. Epidemiology and clinical features of COVID-19: A review of current literature. J Clin Virol. 2020;127:104357.
Crossref - Hong KH, Lee SW, Kim TS, et al. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40(5):351-360.
Crossref - Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev. 2020;296(1):205-219.
Crossref - Choudhary S, Sreenivasulu K, Mitra P, Misra S, Sharma P. Role of Genetic Variants and Gene Expression in the Susceptibility and Severity of COVID-19. Ann Lab Med. 2021;41(2):129-138.
Crossref - Liu X, Zhou H, Zhou Y, et al. Risk factors associated with disease severity and length of hospital stay in COVID-19 patients. J Infect. 2020;81(1):e95-e97.
Crossref - Lacoma A, Mateo L, Blanco I, et al. Impact of Host Genetics and Biological Response Modifiers on Respiratory Tract Infections. Front Immunol. 2019;10:1013.
Crossref - Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Crossref - Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging. 2020;12(11):10087-10098.
Crossref - Benetti E, Tita R, Spiga O, et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 2020;28(11):1602-1614.
Crossref - Camiolo M, Gauthier M, Kaminski N, Ray A, Wenzel SE. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J Allergy Clin Immunol. 2020;146(2):315-324.e7.
Crossref - Mao Q, Chu S, Shapiro S, Bliss JM, De Paepe ME. Increased placental expression of angiotensin-converting enzyme 2, the receptor of SARS-CoV-2, associated with hypoxia in twin anemia-polycythemia sequence (TAPS). Placenta. 2021;105:7-13.
Crossref - Palazzuoli A, Mancone M, De Ferrari GM, et al. Antecedent Administration of Angiotensin-Converting Enzyme Inhibitors or Angiotensin II Receptor Antagonists and Survival After Hospitalization for COVID-19 Syndrome. J Am Heart Assoc. 2020;9(22):e017364.
Crossref - Tang Q, Wang Y, Ou L, et al. Downregulation of ACE2 expression by SARS-CoV-2 worsens the prognosis of KIRC and KIRP patients via metabolism and immunoregulation. Int J Biol Sci. 2021 ;17(8):1925-1939.
Crossref - Srivastava A, Pandey RK, Singh PP, et al. Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations. PLoS One. 2020;15(9):e0238255.
Crossref - Wallentin L, Lindback J, Eriksson N, et al. Angiotensin-converting enzyme 2 (ACE2) levels in relation to risk factors for COVID-19 in two large cohorts of patients with atrial fibrillation. Eur Heart J. 2020;41(41):4037-4046.
Crossref - Azevedo MLV, Zanchettin AC, Vaz de Paula CB, et al. Lung Neutrophilic Recruitment and IL-8/IL-17A Tissue Expression in COVID-19. Front Immunol. 2021;12:656350.
Crossref - D’Alessandro A, Thomas T, Dzieciatkowska M, et al. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J Proteome Res. 2020;19(11):4417-4427.
Crossref - Della-Torre E, Lanzillotta M, Campochiaro C, et al. Respiratory Impairment Predicts Response to IL-1 and IL-6 Blockade in COVID-19 Patients With Severe Pneumonia and Hyper-Inflammation. Front Immunol. 2021;12:675678.
Crossref - Galvan-Roman JM, Rodriguez-Garcia SC, Roy-Vallejo E, et al. IL-6 serum levels predict severity and response to tocilizumab in COVID-19: An observational study. J Allergy Clin Immunol. 2021;147(1):72-80.
Crossref - Sadeghi A, Tahmasebi S, Mahmood A, et al. Th17 and Treg cells function in SARS-CoV2 patients compared with healthy controls. J Cell Physiol. 2021;236(4):2829-2839.
Crossref - Vultaggio A, Vivarelli E, Virgili G, et al. Prompt Predicting of Early Clinical Deterioration of Moderate-to-Severe COVID-19 Patients:Usefulness of a Combined Score Using IL-6 in a Preliminary Study. J Allergy Clin Immunol Pract. 2020;8(8):2575-2581.e2.
Crossref - Wang M, Zhu Q, Fu J, Liu L, Xiao M, Du Y. Differences of inflammatory and non-inflammatory indicators in Coronavirus disease-19 (COVID-19) with different severity. Infect Genet Evol. 2020;85:104511.
Crossref - Xia G, Fan D, Ma C, et al. Hyper-Inflammatory Response Involves in Cardiac Injury Among Patients With Coronavirus Disease 2019. Am J Med Sci. 2021;361(6):718-724.
Crossref - Yuan Y, Wang QP, Sun D, et al. Differences in Immune Responses between Children and Adults with COVID-19. Curr Med Sci. 2021;41(1):58-61.
Crossref - Hoiland RL, Fergusson NA, Mitra AR, et al. The association of ABO blood group with indices of disease severity and multiorgan dysfunction in COVID-19. Blood Adv. 2020;4(20):4981-4989.
Crossref - Niles JK, Karnes HE, Dlott JS, Kaufman HW. Association of ABO/Rh with SARS-CoV-2 positivity: The role of race and ethnicity in a female cohort. Am J Hematol. 2021;96(1):E23-E26.
Crossref - Nauffal V, Achanta A, Goldhaber SZ, Piazza G. Association of ABO blood group type with cardiovascular events in COVID-19. J Thromb Thrombolysis. 2021;51(3):584-586.
Crossref - Sardu C, Marfella R, Maggi P, et al. Implications of AB0 blood group in hypertensive patients with covid-19. BMC Cardiovasc Disord. 2020;20(1):373.
Crossref - Correale P, Mutti L, Pentimalli F, et al. HLA-B*44 and C*01 Prevalence Correlates with Covid19 Spreading across Italy. Int J Mol Sci. 2020;21(15):5205.
Crossref - Lorente L, Martin MM, Franco A, et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Medicina Intensiva. 2021;45(2):96-103.
Crossref - Young BE, Fong SW, Chan YH, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response:an observational cohort study. Lancet. 2020;396(10251):603-611.
Crossref - Jin M, Shi N, Wang M, et al. CD45: A critical regulator in immune cells to predict severe and non-severe COVID-19 patients. Aging. 2020;12(20):19867-19879.
Crossref - Liu F, Long X, Zhang B, Zhang W, Chen X, Zhang Z. ACE2 Expression in Pancreas May Cause Pancreatic Damage After SARS-CoV-2 Infection. Clin Gastroenterol Hepatol. 2020;18(9):2128-2130.e2.
Crossref - Fu L, Wang B, Yuan T, et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis. J Infect. 2020;80(6):656-665.
Crossref - Della-Torre E, Campochiaro C, Cavalli G, et al. Response to: ‘More evidences on which biologic and which pathway is key in severe-critical COVID-19 pneumonia’ by Ferraccioli. Ann Rheum Dis. 2022;81(9):e158.
Crossref - Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J. 2020;34(7):8787-8795.
Crossref - Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631-637.
Crossref - Vargas-Alarcon G, Posadas-Sanchez R, Ramirez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. 2020;260:118313.
Crossref - Lucas JM, Heinlein C, Kim T, et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4(11):1310-1325.
Crossref - Othman H, Bouslama Z, Brandenburg JT, et al. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem Biophys Res Commun. 2020;527(3):702-708.
Crossref - Sanderson SC, Kumari M, Brunner EJ, et al. Association between IL6 gene variants -174G>C and -572G>C and serum IL-6 levels: interactions with social position in the Whitehall II cohort. Atherosclerosis. 2009;204(2):459-464.
Crossref - Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodriguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62-75.
Crossref - Mangalmurti N, Hunter CA. Cytokine Storms:Understanding COVID-19. Immunity. 2020;53(1):19-25.
Crossref - Abdollahi A, Mahmoudi-Aliabadi M, Mehrtash V, Jafarzadeh B, Salehi M. The Novel Coronavirus SARS-CoV-2 Vulnerability Association with ABO/Rh Blood Types. Iran J Pathol. 2020;15(3):156-160.
Crossref - Cooling L. Blood Groups in Infection and Host Susceptibility. Clin Microbiol Rev. 2015;28(3):801-870.
Crossref - Ahluwalia JK, Hariharan M, Bargaje R, Pillai B, Brahmachari V. Incomplete penetrance and variable expressivity: is there a microRNA connection?. Bioessays. 2009;31(9):981-992.
Crossref - Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061-1069.
Crossref - Amoroso A, Magistroni P, Vespasiano F, et al. HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity. Transplantation. 2021;105(1):193-200.
Crossref - Wu Y, Feng Z, Li P, Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin Chim Acta. 2020;509:220-223.
Crossref - Shkurnikov M, Nersisyan S, Jankevic T, et al. Association of HLA Class I Genotypes With Severity of Coronavirus Disease-19. Front Immunol. 2021;12:641900.
Crossref - Shamsi A, Mohammad T, Anwar S, et al. Potential drug targets of SARS-CoV-2: From genomics to therapeutics. Int J Biol Macromol. 2021;177:1-9.
Crossref - Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol J. 2020;17(1):131.
Crossref - Al Barashdi MA, Ali A, McMullin MF, Mills K. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J Clin Pathol. 2021;74(9):548-552.
Crossref - Alon D, Paitan Y, Robinson E, et al. Downregulation of CD45 Signaling in COVID-19 Patients Is Reversed by C24D, a Novel CD45 Targeting Peptide. Front Med (Lausanne). 2021;8:675963.
Crossref - Tee M, Rasli A, Toh JSSK, Abas IH, Zhou F, Liew CS. A Delphi method on the positive impact of COVID-19 on higher education institutions: Perceptions of academics from Malaysia. Front Psychol. 2022;13:1013974.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.