ISSN: 0973-7510
E-ISSN: 2581-690X
Infectious keratitis continues to be a prominent cause of vision impairment worldwide through a variety of causes. Pseudomonas aeruginosa is a Gram-negative bacterium that frequently causes vision-threatening microbial keratitis. P. aeruginosa contains a diverse array of virulence factors, including exoA, exoS, nan1, and lasB, some of which may contribute to its pathogenicity. Because the clinical characteristics of bacterial keratitis vary, making a quick differential diagnosis can be difficult, resulting in a delay in diagnosis and worse outcome. In this study, we performed multiplex polymerase chain reaction to detect the presence of nan1, toxA, exoS, and lasB, and determine their association with distinct clinical presentations of P. aeruginosa-related keratitis. We also performed antibiotic susceptibility testing of the isolates. A total of 49 P. aeruginosa strains were obtained from individuals with keratitis between May 2021 and December 2021 from the Research Institute of Ophthalmology, Giza, Egypt. Results showed that lasB was most expressed gene (81.8%), followed by tox (63.6%) and exoS (31.8%), whereas nan1 was the least commonly expressed gene 1316 (22.7%). The antibiotic susceptibility profile showed that TOB was the least sensitive antibiotic (26.5%), followed by CIP (34.7%), whereas CT was the most sensitive antibiotic (89.8%), followed by GAT (83.7%) and PB (81.6%). Several virulence genes were identified in P. aeruginosa isolates, suggesting that these genes are associated with varying degrees of intrinsic virulence and pathogenicity. Substantial associations between specific virulence genes and the source of infection imply that infection control measures can aid in regulating the distribution of virulence genes among P. aeruginosa strains.
Pseudomonas aeruginosa, Virulence Genes, Keratitis
Pseudomonas aeruginosa is the most frequently isolated Gram-negative bacterium associated with keratitis and is frequently reported as the first or second most prevalent bacterial pathogen linked to corneal infections.1 P. aeruginosa keratitis is a bacterial infection of the cornea that causes blindness globally. Additionally, it accounts for a disproportionate amount of morbidity and mortality associated with bacterial keratitis (BK). Acute ocular infections caused by Pseudomonas typically manifest within 24 h of contact.2
While many researchers agree that disturbance in the corneal epithelium as a result of trauma or use of contact lenses has a role in the development of BK,3 factors underlying rapid infection onset and extensive ulceration continue to be a focus of active investigation. Pseudomonas keratitis is a severe ocular infection that, if not treated aggressively and appropriately, can result in corneal scarring and severe visual impairment.4
P. aeruginosa is an opportunistic pathogen that can infect nearly any tissue, is capable of infecting immunocompromised patients, and is the causative agent of hospital-acquired infections. This bacterium requires minimal nutritional support and is tolerant to a wide range of physical conditions.5
The pathogenesis of P. aeruginosa keratitis is ascribed to several virulence factors in response to specific environmental conditions. The virulence of P. aeruginosa in various eye diseases is associated with a variety of virulence factors that contribute to its pathogenicity, including exoS, nan1, exoA, and lasB.6
Furthermore, P. aeruginosa possesses a remarkable diverse of virulence components, both cell-associated and extracellular, that contribute to its pathogenicity, such as type IV pili, which serve as the principal bacterial adhesion factor, and type III secretion system, which produces exotoxins. When the bacterium attaches to a host cell, the type III secretion system allows it to inject toxins directly into the host cell, disrupting its defense and communication systems.7 P. aeruginosa has been identified to secrete four type III effectors; few, if any, strains secrete all four effectors.8
Exoenzyme S, expressed by exoS, is an ADP ribosyl transferase released directly into the cytoplasm of epithelial cells via the type-III secretion pathway.9 The toxA encodes exoA, which suppresses protein synthesis. The lasB elastase is a zinc metalloprotease expressed by lasB, and it has an elastolytic action against lung tissue.10 Additionally, nan1 encodes a sialidase enzyme required for adhesion to the respiratory tract. Although extracellular neuraminidase is believed to play a critical role in bacterial implantation, the genetic basis of this process remains unclear.11
According to epidemiological studies worldwide, the primary challenge is the spread of resistant and extremely virulent diseases. Owing to the high degree of rRNA conservation throughout the domain Bacteria, chromosomal DNA restriction fragment length polymorphism of rRNA genes (ribotyping) is an excellent tool for strain discrimination both between and within species.12
The conventional drug therapies currently available for P. aeruginosa keratitis are antibiotics, which contribute to bacterial elimination.13 However, the resistance of P. aeruginosa to antibiotics used in clinical practice is increasing via intrinsic and acquired mechanisms. Thus, additional insights into the virulence factors of P. aeruginosa are needed, as the severity of keratitis and its sensitivity to medication depend on the virulence factors of the pathogen.14
This study aimed to detect toxA, exoS, nan1, and lasB in P. aeruginosa ocular keratitis strains using multiplex PCR and determine the association between virulence factors and distinct clinical presentations of P. aeruginosa-related keratitis. Furthermore, we aimed to screen the antibiotic susceptibility profiles of the isolates.
A total of 49 clinical, ocular P. aeruginosa isolates were collected from 49 patients diagnosed with keratitis between May 2021 and December 2021 from the Research Institute of Ophthalmology, Giza, Egypt.
Demographic information, such as name, age, sex, clinical history, and associated results, were collected using a typical clinical history form. Thirty men and 19 women aged 25–65 years were included in this study. Clinical microbiological specimens were collected 48 h prior to initiating antibiotic therapy as shown in table 2.
Table (1):
Specific primers for exoS, toxA, nan1 and lasB genes.
Amplified gene |
Specific Primer |
Amplified region |
|---|---|---|
LasB |
lasf 5’ GGA ATG AAC GAA GCG TTC TC 3’ las r 5’ GGT CCA GTA GTA GCG GTT GG 3’ |
300bp |
ToxA |
toxf 5’ GGT AAC CAG CTC AGC CAC AT 3’ tox r 5’ TGA TGT CCA GGT CAT GCT TC 3’ |
352bp |
ExoS |
exo f 5’ CTT GAA GGG ACT CGA CAA GG 3’ exo r 5’ TTC AGG TCC GCG TAG TGA AT 3’ |
504bp |
Nan1 |
nan f 5’ AGG ATG AAT ACT TAT TTT GAT 3’ nan r 5’TCA CTA AAT CCA TCT CTG ACC CGA TA |
1316bp |
Each patient was examined thoroughly by an ophthalmologist using the slit-lamp biomicroscopic examination. Following thorough eye examination, corneal scrapings were aseptically obtained from the active border of the corneal ulcer using a sterilized platinum Kimura spatula. Isolated specimens were spread on a glass slide for Gram staining and identification, depending on the colony morphology on Nutrient agar and Blood agar (one incubated aerobically and the other anaerobically). Two Chocolate agar plates (one incubated aerobically and the other anaerobically using a gas pack system) and MacConkey agar plates were incubated at 37°C, and isolates were identified based on colony morphology agar and were confirmed using IMViC biochemical tests. Plates that were incubated aerobically were examined for growth after 24 h. Meanwhile, plates that were incubated anaerobically were kept in a closed anaerobic jar for 48 h, the jar was opened thereafter, and the plates were examined for growth. For any negative results from patients not receiving antibiotic treatment, re-scraping was performed during their second visit, and the procedures were repeated.
Multiplex PCR Cycling Protocol
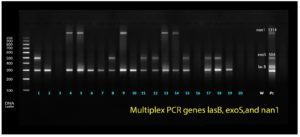
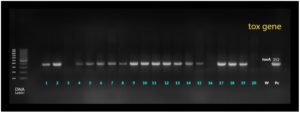
Identification and detection of P. aeruginosa virulence genes nan1, exoS, and lasB were performed via multiplex PCR as shown in Figure 1. Meanwhile, toxA was detected using simplex PCR at different annealing temperatures. The Qiagen multiplex PCR kit (Cat. no. 206145) was used for both protocols as shown in Figure 2. The PCR cocktail contained 2× QIAGEN Multiplex PCR Master Mix, 25 μl 1x10x primer mixture, 2 mM each primer 5.2 l 0.2 μM, 10 μl 1× RNase-free water, and the template DNA, with a total reaction volume of 50 μl. Cycling parameters were as follows: 15 min at 95°C, which activates the HotStarTaq DNA Polymerase, followed by a three-step cycling sequence (denaturation, annealing, and extension). The amplified products were detected using gel electrophoresis. A known P. aeruginosa isolate resistant to all virulence genes was used as the positive control, whereas water was used as the negative control.
Sequence of Primers Used
Detection of the virulence genes was performed using multiplex PCR. Bacterial DNA was extracted using the boiling method. Two to three pure colonies of P. aeruginosa isolates were suspended in 50 µL of distilled water and then vortexed to homogenate the suspension. Subsequently, the suspension was incubated at 94°C for 10 min and then centrifuged. The supernatant was collected in a clean tube and stored at −20°C as a template DNA stock.
Table (2):
Mean, standard deviation (SD), frequencies (n), percentages and results of Student’s t-test and Chi-square test for comparisons between base line characteristics in the two groups.
| Base line characteristics | Non-MDR (n = 27) |
MDR (n = 22) | P-value |
|---|---|---|---|
| Age (Years) | 0.148 | ||
| Mean (SD) | 42.1 (12.4) | 36.5 (14.5) | |
| Gender [n (%)] | 0.367 | ||
| Male | 15 (55.6%) | 15 (68.2%) | |
| Female | 12 (44.4%) | 7 (31.8%) | |
PCR was performed using specific primers for the virulence genes.15 The final reaction mixture contained 25 μl of (Taq polymerase, reaction buffer, MgCl, and dNTPs), 2 μl of each 10 pmol forward and reverse specific primers, 5 μl of the DNA template, and 13 μl of (ddH2O) as shown in table 1, for a total volume of 25 L. A thermal cycler (Techne, England) was used to cycle the reaction tubes. The PCR conditions were as follows: 5 min at 94°C (denaturation), followed by 35 cycles of denaturation (1 min, 94°C) and annealing (55°C for toxA and 58°C for exoS, nan1, and lasB). PCR products were separated via gel electrophoresis using 2% agarose gel containing 0.5 µg/ml ethidium bromide.
Data Analysis
All statistical analyses were performed using SPSS version 22.0 for Windows (SPSS Inc., Chicago, IL, USA).
Sex and Age Ratio
P. aeruginosa was found in 30/49 (61.2%) men and 19/49 (38.8%) women. Prevalence was higher in patients aged 55–65 years, which included 27 of the cases (55%).
Multiplex PCR Detection for Virulence Genes
Detection of virulence genes has been limited to multidrug-resistant (MDR) bacteria, which are increasing at an alarming rate worldwide, thus causing serious health problems. The incidence of infections due to MDR bacteria has been accompanied by poor outcomes up to eye loss. Therefore, overcoming bacterial resistance has become an urgent and unmet challenge that should be properly addressed to determine a line of treatment, leading to prevention of poor clinical outcomes in comparison with the non-MDR bacteria. Among the isolates, the most expressed gene was lasB (81.8%), followed by toxA (63.6%) and exoS (31.8%). Meanwhile, nan1 (22.7%) was the least commonly expressed gene 1316 as shown in table 3.
Table (3):
Percentages of virulence genes in MDR group.
Gene |
N (22) |
% |
|---|---|---|
LasB |
18 |
81.8 |
ExoS |
7 |
31.8 |
Nan1 |
5 |
22.7 |
ToxA |
14 |
63.6 |
Antibiotic Susceptibility Testing
Antibiotic susceptibility profiling was performed using the Kirby Bauer disc diffusion method using anti-pseudomonal antimicrobial agents often used in clinical practice to treat keratitis. All antibiotic discs listed in Table (4) were supplied by Oxoid (Basingstoke, UK) and the results were interpreted following the CLSI 2018 guidelines.16
Table (4):
Percentages of sensitive antibiotic susceptibility test arranged from lowest to highest.
Antibiotic type |
N (49) |
% |
|---|---|---|
Tobramycin (TOB) |
13 |
26.5 |
Ciprofloxacin (CIP) |
17 |
34.7 |
Gentamycin (GEN) |
19 |
38.8 |
Amikacin (AK) |
20 |
40.8 |
Chloramphenicol (C) |
22 |
44.9 |
Meropenem (MEM) |
24 |
49.0 |
Ofloxacin (OFX) |
25 |
51.0 |
Levofloxacin (LEV) |
26 |
53.1 |
Cefazolin (CZ) |
28 |
57.1 |
Imipenem (IPM) |
30 |
61.2 |
Ceftazidime (CZA) |
30 |
61.2 |
Azithromycin (AZM) |
31 |
63.3 |
Pipracillin-Tazobactam (PIT) |
33 |
67.3 |
Ceftazidime-Avibactum (CZA-AV) |
38 |
77.6 |
Polymyxin B (PB) |
40 |
81.6 |
Gatifloxacin (GAT) |
41 |
83.7 |
Colistin (CT) |
44 |
89.8 |
MDR bacteria are described as those that are resistant to at least one agent in three antibiotic classes (e.g. beta-lactam, aminoglycoside, macrolide).6 Results showed that the least sensitive antibiotic was TOB (26.5%), followed by CIP (34.7%), while the most sensitive antibiotic was CT (89.8%), followed by GAT (83.7%) and PB (81.6%).
A total of 25 cases showed ulcers with abscess formation 8 (29.6%) in non MDR isolates and 17 (77.2%) in MDR isolates, showing the highest prevalence. Meanwhile, all 49 patients showed unilateral corneal ulceration (100%) (P <0.005, significant) as shown in table 5.
Table (5):
Distribution of clinical manifestations seen among MDR and non-MDR groups.
Clinical Manifestation |
Non-MDR (n = 27) |
MDR (n = 22) |
|---|---|---|
Ulcer with Redness |
15 (55.5%) |
2 (9%) |
Ulcer with Severe lid edema |
4 (14.8%) |
3 (13.6%) |
Ulcer with abscess |
8 (29.6%) |
17 (77.2%) |
If not treated promptly and appropriately, microbial keratitis caused by Pseudomonas aeruginosa17 causes significant ocular morbidity and may result in blindness.18
P. aeruginosa is a known opportunistic human pathogen that is capable of infection via attachment, colonization, local invasion, and dissemination, leading to systemic illness. Pseudomonal keratitis frequently manifests as ulceration of the stroma with underlying epithelial deficiency, resulting in severe and frequent rapid stromal melting, which may lead to corneal perforation and vision loss.19
Characterization of P. aeruginosa virulence factors is critical in understanding its pathophysiology and aids in the development of novel antimicrobial treatments against MDR strains.20
In our study, we reported that the incidence of P. aeruginosa infection in male and female patients were 61.2% and 38.8%, respectively, which is consistent with a previous study.21 O’Callaghan et al.21 found that male patients had a greater incidence of P. aeruginosa infection (56/XX, 64.36%) than female patients (31/XX, 35.63%). Usually, men engage in routine outdoor work and are thus commonly exposed to infectious settings.22
In our study, we observed that P. aeruginosa infection was more prevalent in patients aged 55–65 years, accounting for 55% of cases. This finding is in accordance with a study conducted by Eichenberger et al.22 who noted an incidence rate of 41.37% in patients aged 60–79 years. These results indicate P. aeruginosa infection is more prevalent in older patients. This could be explained by a decline in the function of the immune system and an extended stay of hospitalization.
Furthermore, we observed that the most common clinical sign was ulcer associated with abscess formation, in 8 patients (29.6%) in the non MDR group and 17 (77.2%) in the MDR group, followed by ulcers with redness and edema in 15 (55.5%) in the non MDR group and 2(9%)in the MDR group. Ulcers with lid edema had the highest curability rate, whereas ulcers with abscess had lowest curability rate, which is attributed to P. aeruginosa virulence factors involved not only in the early induction of the epidermis inflammatory response, but also in bacterial invasion and cutaneous persistence.23 The high presentation of ulcers associated with abscesses in our study population may be due to late patient visits that cause ulcer deterioration. Most of the previous studies did not report the analysis of clinical signs, as no distinctive or exclusive signs that could help in the identification of the causative organisms were observed. However, clinical experience may lead to a probable etiological diagnosis in some cases.
In addition, we reported the prevalence of MDR isolates in 22 out of 49 cases, giving a prevalence of 44.9%, which was similar to a previous study that observed 23 (50%) MDR isolates.25 Additionally, the increased prevalence of MDR P. aeruginosa was likely due to an impairment of the national antibiotic management policy, and easy access to medicines without a physician’s prescription may have led to an increase in MDR isolates. As expected, MDR-PA was significantly associated with poor visual performance and prognosis.
For the antibiotic susceptibility testing, Manandhar et al.26 and Naik et al.27 reported similar results in antibiogram. The least sensitive antibiotic was TOB (26.5%) followed by CIP (34.7%) while the highest positive sensitivity was found with CT (89.8%) followed by GAT (83.7%) and PB (81.6%)
The pathogenicity of P. aeruginosa is complex. The virulence gene lasB is a key protease of P. aeruginosa. Of all the isolates investigated, 81.8% harbored lasB. This study confirmed the findings of previous studies. The presence of the lasB in all environmental and clinical strains demonstrates its critical role in the survival of P. aeruginosa in a variety of environments. P. aeruginosa isolates typically exhibit cytotoxic or invasive characteristics that are known to involve exoS (encoding exotoxin S) or exoU (encoding exotoxin U).
P. aeruginosa is the most common pathogen causing vision-threatening corneal infections. Numerous virulence genes identified in P. aeruginosa isolates have shown that they confer various degrees of intrinsic virulence and pathogenicity that may have various effects on the progression of infection. This study provides additional insight into the pathophysiology of Pseudomonas keratitis and may aid in the development of novel antimicrobial drugs, as a result of the increased prevalence of MDR-PA and poor clinical outcomes.
ACKNOWLEDGMENTS
The authors would like to thank members of Research Institute of Ophthalmology for their support to carry out this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All database generated or analyzed during this study are included in the manscript.
- Austin A, Lietman T, Rose-Nussbaumer J. Update on the Management of Infectious Keratitis. Ophthalmology. 2017;124(11):1678-1689.
Crossref - Bahl CD, St Laurent JD, Karthikeyan RS, et al. The cif Virulence Factor Gene Is Present in Isolates from Patients with Pseudomonas aeruginosa Keratitis. Cornea. 2017;36(3):358-362.
Crossref - Zimmerman AB, Nixon AD, Rueff EM. Contact lens associated microbial keratitis: practical considerations for the optometrist. Clin Optom (Auckl). 2016;8:1-12.
Crossref - Ting DSJ, Ho CS, Deshmukh R, Said DG, Dua HS. Infectious keratitis: an update on epidemiology, causative microorganisms, risk factors, and antimicrobial resistance. Eye (London). 2021;35(4):1084-1101.
Crossref - Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39.
Crossref - Kandasamy K, Thirumalmuthu K, Prajna NV, Lalitha P, Mohankumar V, Devarajan B. Comparative genomics of ocular Pseudomonas aeruginosa strains from keratitis patients with different clinical outcomes. Genomics. 2020;112(6):4769-4776.
Crossref - Jurado-Martin I, Sainz-Mejias M, McClean S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int J Mol Sci. 2021;22(6):3128.
Crossref - Azimi S, Kafil HS, Baghi HB, et al. Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect Control. 2016;11:Doc04.
Crossref - Kroken AR, Chen CK, Evans DJ, Yahr TL, S Fleiszig MJ. The Impact of ExoS on Pseudomonas aeruginosa Internalization by Epithelial Cells Is Independent of fleQ and Correlates with Bistability of Type Three Secretion System Gene Expression. mBio. 2018;9(3):e00668-18.
Crossref - Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154(6):1570-1583.
Crossref - Bogiel T, Prazynska M, Kwiecinska-Pirog J, et al. Carbapenem-Resistant Pseudomonas aeruginosa Strains-Distribution of the Essential Enzymatic Virulence Factors Genes. Antibiotics (Basel). 2020;10(1):8.
Crossref - Nikbin VS, Aslani MM, Sharafi Z, Hashemipour M, Shahcheraghi F, Ebrahimipour GH. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran J Microbiol. 2012;4(3):118-123. PMCID: PMC3465536
- Proctor LL, Ward WL, Roggy CS, et al. Potential Therapeutic Targets for Combination Antibody Therapy against Pseudomonas aeruginosa Infections. Antibiotics (Basel). 2021;14;10(12):1530.
Crossref - Zheng P, Renee R, Bernard R, et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Mitov I, Strateva T, Markova B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of pseudomonas aeruginosa. Braz J Microbiol. 2010;41(3):588-595.
Crossref - Clinical and Laboratory Standards Institute (CLSI). 2019. https://clsi.org/media/2663/m100ed29_sample.pdf
- Behzadi P, Barath Z, Gajdacs M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug- Resistant Pseudomonas aeruginosa. Antibiotics. 2021;10(1):42.
Crossref - Ung L, Bispo PJM, Shanbhag SS, et al. The persistent dilemma of microbial keratitis: Global burden, diagnosis, and antimicrobial resistance. Surv Ophthalmol. 2019;64(3):255-271.
Crossref - Miller D. Pharmacological treatment for infectious corneal ulcers. Expert Opin Pharmacother. 2013;14(5):543-560.
Crossref - Bazghandi SA, Arzanlou M, Peeridogaheh H, Vaez H, Sahebkar A, et al. Prevalence of Virulence Genes and Drug Resistance Profiles of Pseudomonas aeruginosa Isolated from Clinical Specimens. Jundishapur J Microbiol. 2021;14(8):e118452.
Crossref - O’Callaghan R, Caballero A, Tang A, Bierdeman M. Pseudomonas aeruginosa Keratitis: Protease IV and PASP as Corneal Virulence Mediators. Microorganisms. 2019;7(9):281.
Crossref - Eichenberger EM, Thaden JT. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics (Basel). 2019;8(2):37.
Crossref - Charles D, Eric D, Jeff G, Levesque RC, Lau GW. An Organ System-Based Synopsis of Pseudomonas aeruginosa Virulence. Virulence. 2021;12:(1)1469-1507.
Crossref - Borkar DS, Fleiszig SM, Leong C, et al. Association between cytotoxic and invasive Pseudomonas aeruginosa and clinical outcomes in bacterial keratitis. JAMA Ophthalmol. 2013;131(2):147-153.
Crossref - Gill JS, Arora S, Khanna SP, Kumar HKVS. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level Intensive Care Unit. J Global Infect Dis. 2016;8(4):155-159.
Crossref - Manandhar S, Adhikari S, Rajbhandari S. Phenotypic assays for detection of AmpC and MBL producers among the clinical isolates of multi drug resistant Pseudomonas aeruginosa. Tribhuvan University Journal of Microbiology. 2018;4(1):23-31.
Crossref - Naik P, Pandey S, Gagan S, Biswas S, Joseph J. Virulence factors in multidrug (MDR) and Pan-drug resistant (XDR) Pseudomonas aeruginosa: a cross-sectional study of isolates recovered from ocular infections in a high-incidence setting in southern India. J Ophthal Inflamm Infect. 2021;11(1):36.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.