ISSN: 0973-7510
E-ISSN: 2581-690X
To produce an active metabolite of Chlordiazepoxide by fungal biotransformation in an easy and economic way and also to develop microbial models for drug metabolism studies. Chlordiazepoxide is metabolized in the liver by CYP3A4 and forms major active metabolite N-desmethyl chlordiazepoxide. The focus of the study was to explore the ability of six distinct fungi to biotransform the drug Chlordiazepoxide to its metabolites. Induction, Inhibition and kinetic studies were also conducted to find out the type and capability of enzyme involved in fungal biotransformation. The screening studies were performed and fermentation protocol was designed with two controls (culture control and drug control) and one sample. Extract metabolite samples were reconstituted and analysed using HPLC. Induction, Inhibition studies were conducted similarly by maintaining its respective controls using CYP3A4 inducer (Carbamazepine) and inhibitor (Fluoxetine), further kinetic studies were performed to find Km and Vmax of fungal biotransformation of Chlordiazepoxide. Among six organisms Aspergillus ochreus has shown an extra peak at 6.9 min. in HPLC when compared with its controls indicated the formation of metabolite. The metabolite thus formed was identified, isolated and structure was confirmed by mass spectrometry and NMR spectroscopy as Nor-chlordiazepoxide. During inhibition and induction studies, it was found that quantity of the metabolite was increased with inducer and decreased with inhibitor. The Km and Vmax of fungal metabolism of Chlordiazepoxide was 1.928 µg/ml and 0.1802 µg/ml/hr respectively. Aspergillus ochreus has the ability to biotransform the Chlordiazepoxide to its active metabolite by CYP3A4 like enzyme and it followed Michaelis-Menten kinetics.
Chlordiazepoxide, Fungal Transformation, Aspergillus ochreus, Michaelis Menten Kinetics, Induction Studies and Inhibition Studies
Chlordiazepoxide which is chemically known as 7-chloro-2-methylamino-5-phenyl-3H-1,4 benzodiazepine-4-oxide and it is an anti epileptic drug. Metabolism Chlordiazepoxide is metabolised in liver via N-demethylation and results in formation of an active metabolite N-desmethyl chlordiazepoxide (Norchlordiazepoxide). It undergoes deamination to form the demoxepam, and demethylation to form Norchlordiazepoxide, this step is very rapid in metabolism of chlordiazepoxide.1
Chlordiazepoxide was selected in the current study because, it was evident that active metabolite, N-desmethyl chlordiazepoxide has supreme pharmacological activity than its parent drug in mammals. Chlordiazepoxide is a substrate for CYP3A4 in mammals. Fungal metabolism studies were proved as potential suitable methods because of their inherent enzymatic systems which can produce variety of chemical compounds when compared to synthetic procedures.2-17 In this context, fungal biotransformation studies are essential to produce active metabolite of chlordiazepoxide by fungi and also to develop microbial model for 3A4 enzyme. Hence, in present study, screening of fungi for their ability to biotransform chlordiazepoxide was conducted to identify the fungi which could be able to biotransform the chlordiazepoxide. Then, nature of enzyme in the fungi was determined by metabolism induction and inhibition studies. The affinity of the fungal enzymes towards drug together with the effect of concentration of substrate drug and incubation time on percentage of metabolite formation by the fungi was estimated by enzyme kinetic studies.
Fungi
Aspergillus niger (NCIM-589), Aspergillus fumigatus (NCIM-902), Aspergillus ochreus (NCIM-1140) Cunnighamella elegans (NCIM-689), Cunnighamella blackesleeana(NCIM-691), Cunnighamella echinulata (NCIM-687), were obtained from National chemical laboratory(NCL) Pune, India.
Chemicals
Chlordiazepoxide was a gift sample from Lake Chemicals, Bangalore. Remaining chemicals were procured from S.D fine chemicals. The solvents used for analysis were HPLC grade and obtained from Indian scientific Pvt ltd, Tirupati, A.P.
Stock culture maintenance
All cultures were maintained on agar slants at 4°C18 and viability of all cultures was sustained by frequent sub culturing for every 6 months. The suitable growth medium for fungal cultures is Potato dextrose broth.19 The growth medium was sterilised before use by autoclaving at 121°C for 30 min.20
Fungal screening studies
Screening studies were carried out by keeping three 250 ml Erlenmeyer flask containing 50ml of broth media for drug control, culture control and sample for each culture. Among three flasks, drug alone was added to broth medium without culture to drug control flask. One loopful of respective fungi culture was added to culture control flask without drug addition. Sample flask is added with both drug and culture. All the flasks were subjected to incubation in shaker incubator. (CIS 24Remi instruments, Mumbai, India) for 72 hrs under uniform conditions to attain the adequate growth of fungi for biotransformation of Chlordiazepoxide.21 Then drug and formed metabolites were extracted to perform further analysis in HPLC.
Inhibition and induction studies
In mammals, Chlordiazepoxide was metabolised by CYP 3A4 enzyme.22 The present study was concentrated to conduct enzyme inhibition and induction studies with the help of CYP 3A4 inducer (carbamazepine) and inhibitor (fluoxetine) to identify the nature of enzyme accountable for the production of metabolite of Chlordiazepoxide with the confirmed fungus from screening studies. For induction and inhibition studies, 6 flasks, one sample and five controls were set, which were namely Blank I (drug control), Blank II (Culture control) comprises of selected fungus, Blank III (inhibitor/inducer control), Blank IV (inhibitor/inducer and substrate control) and Blank V (Culture +Inhibitor/Inducer control) to inspect the intervention of substrate, inhibitor/inducer in association with the media. Sample for induction and inhibition studies contained drug, inducer/inhibitor and culture. Stock solutions of 1mg/ml concentration of Chlordiazepoxide, CYP3A4 inducer (Carbamazepine) and inhibitor (Fluoxetine) were prepared separately. Assigned flasks containing 50 ml of potato dextrose broth were aseptically transferred with 0.5ml of drug solution, inhibitor solution and inducer solution from prepared stock solutions and fungal culture as per aforementioned protocol. All the flasks were incubated for 72 hrs and contents were subjected to inactivation and extraction. The formed metabolites after extraction were analysed in similar to screening studies. Then, the percentage metabolite formed was calculated with the help of peak areas of metabolite found in HPLC chromatograms.33
Enzyme kinetic studies
The enzyme affinity towards the given substrate drug in the fungal biotransformation was evaluated by these enzyme kinetic studies. The confirmed fungal strain was inoculated in labelled sample flasks consisting different substrate (Chlordiazepoxide) concentrations like 10μg/ml, 20μg/ml, 30μg/ml, 40 μg/ml, 50 μg/ml and 60 μg/ml, and incubated at predetermined conditions. Samples were collected aseptically from each flask at different incubation time intervals like 24,36,48,60 and 72 hrs and each sample extracted for the metabolite and the percentage of metabolite formed at each concentration of substrate and different time points was calculated.24
Extraction method
Samples collected after pre fixed incubation period in all the above studies were heated to inactivate the grown fungi at 50°C for 30 minutes. Then centrifugation was done at 3000 rpm for 10 min for all samples. (Laboratory Centrifuge C-854/8, Remi instruments, Mumbai, India). Supernatant was collected from all tubes and were stored in refrigerator. Diethyl ether was used to treat supernatant to extract Chlordiazepoxide and formed metabolites.25,26 Then organic layer was collected in screw cap bottles and kept for air drying. The dried samples were analysed by HPLC after reconstitution with mobile phase, besides pure drug Chlordiazepoxide was also injected into HPLC to set it as a standard.27
Analytical methods
High performance liquid chromatography28,29
Extracted drug and metabolites were isolated by using a phenomenex luna 5µ C18(2) 100A 250 X 4.60mm (Phenomenex, USA) column and solvent system consisted of Methylene chloride: methanol (0.5:9.5) at a flow rate of 1ml/min in a HPLC system (Shimadzu, Kyoto, Japan) with LC 20 AD pump solvent delivery module and SPD 20AV UV detector. Sensitivity was set at 0.0001aufs. The UV detector wavelength was set at 217 nm. The mobile phase was filtered through a 0.45 μm filter, degassed using sonicator for 15 min. and run time was set for 30 min.
Mass spectrometry and Proton Nuclear Magnetic Resonance spectroscopy
The metabolite found in samples of fungi screened at the time of elution in HPLC analysis was collected. The molar mass of the formed metabolite was analysed through Mass spectrometry. Mass spectrometer (Agilent technologies, Germany) model was API 3000MS operated in the electron spray ionization (ESI) mode. Ionization was carried out in positron mode using ion trap detector (3.5 kV, 325°C, 210 psi). The structural confirmation of metabolite isolated from HPLC was done by 1H NMR spectroscopy by BRUKER AVANCE 500 MHz NMR spectrometer while the solvent used for analysis of Chlordiazepoxide and its metabolite was Deuterated methanol.30
Quantification of metabolite
The quantification of the metabolite formed in terms of percentage was obtained from peak area of metabolite samples collected during induction, inhibition and kinetic studies in relation to the peak area of drug in HPLC analysis.31
Kinetic studies
The data obtained while kinetic studies in terms of percentage metabolite was exported to Micro Soft EXCEL to find out the velocity of reaction by plotting graph between incubation time and percentage metabolite formed at each concentration of substrate from the slope of the graph. Later the velocity obtained for each concentration by above method was applied to the Michaelis–Menten equation using nonlinear regression in Graph Pad Prism 5.0 and best-fit values for Km and Vmax were obtained in terms of correlation coefficients which were serve as indicators to find the extent of affinity of fungal enzyme towards Chlordiazepoxide.32,33
Screening studies
High Performance Liquid chromatography
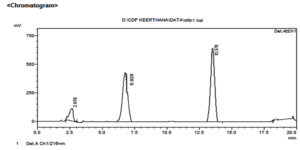
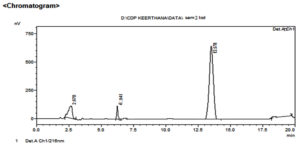
In this proposed research work, six different fungi were screened to study biotransformation of chlordiazepoxide. The results of HPLC analysis data is given in Table 1. HPLC chromatogram of chlordiazepoxide is shown in Figure 1. In HPLC analysis of six fungal cultures, sample of Aspergillus ochreus culture has shown an extra peak at 6.9 min. when compared with its two controls among which blank I (drug control) showed the presence of drug peak at retention time of 13.5 min which was coinciding with drug peak in chromatogram of chlordiazepoxide while no drug peak was observed in chromatogram of the blank II (culture control). The peak at retention time of 2.6 minutes depicted solvent peak found in all the three samples. The extra peak in HPLC chromatogram of sample of Aspergillus ochreus is indicative for formation of metabolite due to the biotransformation of chlordiazepoxide by Aspergillus ochreus and it is represented in Figure 2 and in Table 1. No extra peaks were observed in the samples of other cultures when compared with their controls. Since, this fungus alone metabolised chlordiazepoxide, the elute of extra peak was collected from HPLC and further analysed by Mass and 1H NMR spectroscopy to confirm the metabolite structure and to establish its metabolic pathway in fungi.
Table (1):
The results of HPLC analysis of chlordiazepoxide and its metabolites in different culture extracts.
| Name of the organism | Retention Time (min.) | |||
|---|---|---|---|---|
| Blank I (Drug control) |
Blank II (Culture control) |
Sample | Standard | |
| Cunnighamella elegans (NCIM-689) | 2.6 13.5 |
2.6 | 2.6 13.5 |
13.5 |
| Cunnighamella echinulata(NCIM-687) | 2.6 13.5 |
2.6 | 2.6 13.5 |
13.5 |
| Cunnighamella blackesleeana(NCIM-691) | 2.6 13.5 |
2.6 | 2.6 13.5 |
13.5 |
| Aspergillus niger(NCIM-589) | 2.6 13.5 |
2.6 | 2.6 13.5 |
13.5 |
| Aspergillus fumigatus(NCIM-902) | 2.6 13.5 |
2.6 | 2.6 13.5 |
13.5 |
| Aspergillus ochreus-(NCIM-1140) | 2.6 13.5 |
2.6 | 2.6 6.9*(CDM) 13.5 |
13.5 |
“*” – Metabolite peak.
| Peak Table | |||||
| Detector A Ch1 216nm | |||||
| Peak# | Ret. Time | Area | Height | Area % | Height % |
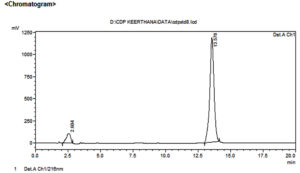
| 1 | 2.604 | 2423229 | 105387 | 8.264 | 8.185 |
| 2 | 13.578 | 26899615 | 1182096 | 91.736 | 91.815 |
| Total | 29322845 | 1287483 | 100.000 | 100.000 | |
Figure 1. HPLC chromatogram of chlordiazepoxide
| Peak Table | |||||
| Detector A Ch1 216nm | |||||
| Peak# | Ret. Time | Area | Height | Area % | Height % |
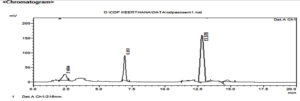
| 1 | 2.604 | 191026 | 30958 | 5.993 | 10.369 |
| 2 | 6.951 | 878208 | 85731 | 27.552 | 28.714 |
| 3 | 13.578 | 7728230 | 8171 | 67.254 | 62.737 |
| Total | 8797644 | 124860 | 100.000 | 100.000 | |
Figure 2. HPLC chromatogram of chlordiazepoxide from culture extracts of Aspergillus ochreus
Mass spectrometry
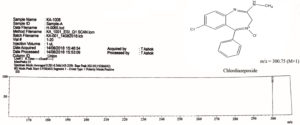
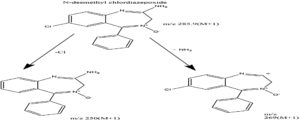
Mass spectrum of drug and metabolite and its fragmentation pattern are shown in Figure 3, 4 and 5. Mass spectrum of chlordiazepoxide was compared with mass spectrum of metabolite CDM. The mass spectrum of drug has shown a molecular ion peak at m/z 300.75 (M+1) which is equal to the molecular weight of chlordiazepoxide as in Figure 3.
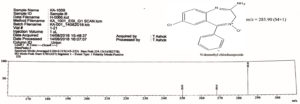
Mass spectrum of CDM, a metabolite produced by Aspergillus ochreus has shown a molecular ion peak at m/z 285.90 [M+1] that is equal to the molecular weight of Desmethyl chlordiazepoxide as shown in Figure 4. The structure of Desmethyl chlordiazepoxide is also supported by its fragment peaks at m/z 250 and 269 as per mass fragmentation pattern shown in Figure 5.
Proton NMR Spectroscopy
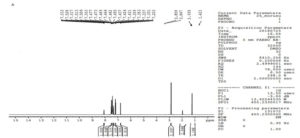
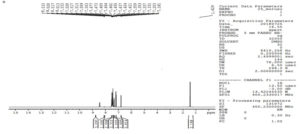
The structure of metabolite, CDM was further confirmed by proton NMR spectroscopy. proton NMR spectrum of chlordiazepoxide is shown in Figure 6 was used to compare with proton NMR spectrum of metabolite.
N-desmethyl chlordiazepoxide, the structure of CDM was confirmed by the absence of a triplet at δ 0.79 in its proton NMR spectrum (Figure 7) stated the loss of methyl group from the structure of the drug, and appearance of a doublet at δ 8.51 represented the presence of NH2 group (possible by loss of methyl group). NMR proton assignment of chlordiazepoxide and its metabolite, CDM is represented in Table 2. Hence, the formation of N-desmethyl chlordiazepoxide, an active metabolite of chlordiazepoxide was confirmed.
Table (2):
NMR proton assignment of chlordiazepoxide and its metabolite, CDM.
Compound Name |
1H NMR proton assignment |
|---|---|
Chlordiazepoxide |
0.79(s,3H,CH3), 2.0(s,1H,OH), 4.42(s,2H,CH2), 6.80(s,1H,H), 7.33-7.46(m,4H, CH), 7.50-7.53(s,1H,CH), 7.97-7.97(d,2H,H ethylene). |
Metabolite, CDM |
2.0(s,1H,OH), 4.42(s,2H,CH2), 6.80(s,1H,H), 7.33-7.46(m,4H, CH), 7.50-7.53(s,1H,CH), 7.97-7.97(d,2H,H ethylene), 8.51(d,2H,NH2) |
* Bold face indicates that additional peaks observed in the respective proton NMR spectra.
Induction and Inhibition studies of fungal metabolism
It was found that, an active metabolite N-desmethyl chlordiazepoxide was formed by Aspergilus ochreus similar to mammals.34 So, microbial metabolism induction and inhibition studies were conducted to identify the nature of enzyme involved in fungal biotransformation of chlordiazepoxide, using CYP 3A4 inducer (carbamazepine) and inhibitor (fluoxetine). The results of HPLC data of chlordiazepoxide and its metabolite (CDM) during induction and inhibition studies by Aspergilus ochreus is represented in Table 3. The percentage of metabolite, CDM was found to be increased in the presence of CYP 3A4 inducer and decreased in the presence of CYP 3A4 inhibitor.
Table (3):
HPLC data of induction and inhibition studies of chlordiazepoxide biotransformation.
| Name of the organism | Type of study | Name of drug | Peak area (mV/s) | Percentage metabolite formed | |
|---|---|---|---|---|---|
| Drug | Metabolite | ||||
| Aspergillus ochreus | Biotransformation study | Chlordiazepoxide | 7728230 | 878208 | 8.8 |
| Induction study | Carbamazepine (as CYP 3A4 inducer) |
9630819 | 992868 | 9.7 | |
| Inhibition study | Fluoxetine (as CYP 3A4 inhibitor) |
8924663 | 22424 | 0.26 | |
| Peak Table | |||||
| Detector A Ch1 216nm | |||||
| Peak# | Ret. Time | Area | Height | Area % | Height % |
| 1 | 2.670 | 2348597 | 108694 | 8.862 | 12.694 |
| 2 | 6.928 | 992868 | 501429 | 10.30 | 31.88 |
| 3 | 13.578 | 9630819 | 962589 | 80.828 | 55.526 |
| Total | 12972284 | 1572712 | 100.00 | 100.00 | |
Figure 8. HPLC Chromatogram of chlordiazepoxide in the presence of carbamazepine from culture extracts of Aspergillus ochreus (CYP3A4 induction study)
| Peak Table | |||||
| Detector A Ch1 216nm | |||||
| Peak# | Ret. Time | Area | Height | Area % | Height % |
| 1 | 2.670 | 2348597 | 108694 | 8.862 | 12.694 |
| 2 | 6.941 | 22424 | 32362 | 0.19 | 0.257 |
| 3 | 13.578 | 8924663 | 12427414 | 90.94 | 87.043 |
| Total | 11295684 | 12568470 | 100.00 | 100.00 | |
Figure 9. HPLC Chromatogram of Chlordiazepoxide in the presence of fluoxetine from culture extracts of Aspergillus ochreus (CYP3A4 inhibition study)
Metabolite-CDM
Percentage of metabolite formation was increased in case of samples incubated with Aspergillus ochreus in presence of CYP 3A4 inducer from 8.8% to 9.7 % (Figure 8) and in presence of CYP 3A4 inhibitor it was decreased to 0.26% (Figure 9). It demonstrated that, CYP 3A4 like enzyme might be involved in the biotransformation of chlordiazepoxide by Aspergillus ochreus.
Enzyme kinetic studies
Enzyme kinetic studies of Chlordiazepoxide metabolism were conducted with Aspergillus ochreus for metabolite, CDM. Six different substrate concentrations (10μg/ml to 60μg/ml) were used in this study. The results of HPLC analysis obtained in the form of peak area were used to calculate percentage metabolite formed at different time points at each concentration. For each concentration of substrate, a graph was plotted between percentages of metabolite v/s incubation time. Slope values obtained from each plot were considered as velocity of metabolic reaction at each concentration.
During enzyme kinetic studies, samples were analyzed by HPLC after collecting at regular incubation time intervals of 24,36,48,64 and 72 hrs. The graphs plotted between percentage of metabolite v/s incubation time were analyzed to find out the effect of incubation time on metabolite formation and velocity of reaction.
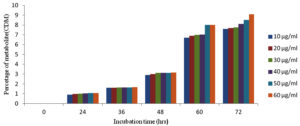
As the incubation time was increased the percentage metabolite (CDM) formation was also increased up to 72 hrs of incubation with all six concentrations of substrate as shown in Figure 10.
Figure 10. Effect of incubation time on percentage metabolite CDM formation with six different concentrations of substrate
Percentage of metabolite CDM formed during kinetic studies of chlordiazepoxide metabolism by Aspergillus ochreus at different incubation time points with six concentrations of substrate represented in Table 4.
Table (4):
Percentage metabolite formed during kinetic studies by Aspergillus ochreus.
| Substrate concentration (µg/ml) | Incubation time (hrs.) | ||||
|---|---|---|---|---|---|
| 24 | 36 | 48 | 60 | 72 | |
| 10 | 0.90 % | 1.60 % | 2.9 % | 6.7 % | 7.6 % |
| 20 | 0.98 % | 1.61 % | 3.0 % | 6.9 % | 7.7 % |
| 30 | 1.01 % | 1.62 % | 3.11 % | 7.0 % | 7.75 % |
| 40 | 1.04 % | 1.62 % | 3.12 % | 7.03 % | 8.1 % |
| 50 | 1.05 % | 1.63 % | 3.13 % | 7.1% | 8.3 % |
| 60 | 1.06 % | 1.65 % | 3.16 % | 7.2 % | 9.1 % |
Slope values obtained from plots drawn between percentages of metabolite (CDM) v/s incubation time, for six different concentrations which were considered as velocity of metabolic reaction by enzyme in Aspergillus ochreus as represented in Table 5.
Table (5):
Slope values for metabolite, CDM with six different concentrations.
Substrate concern. (µg/ml) |
Slope values(m) or velocity of reaction (µg/ml/min) CDM |
|---|---|
10 |
0.113 |
20 |
0.115 |
30 |
0.116 |
40 |
0.119 |
50 |
0.129 |
60 |
0.134 |
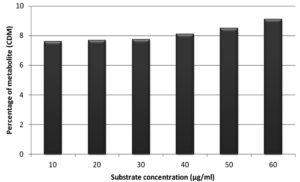
Figure 11. Effect of substrate concentration on percentage of metabolite (CDM) after incubation of 72 hrs
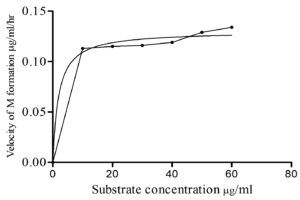
Six different substrate concentrations (10 µg/ml – 60µg/ml) were used to study the influence of substrate concentration on percentage of metabolite formation (CDM) for an incubation of 72 hrs and is outlined in Figure 11. The percentage of formation of CDM was increased with increase in substrate concentration from 10 to 60µg/ml. Then, the velocity of reaction (Table 5) was plotted against substrate concentration (Figure 12) by applying Michaelis-Menten kinetics in Graph Pad Prism 5.0 software to get kinetic parameters Km and Vmax. Velocity of reaction was increased with increase in substrate concentration in case of CDM formation by Aspergillus ochreus.
Screening studies
The ability of fungi to biotransform the drug chlordiazepoxide, a CYP3A4 substrate35 to its active metabolite was investigated using six fungal cultures. The similar enzymatic systems like CYP 450 enzymes important for biotransformation of chlordiazepoxide may be present in fungi, hence, any one of these may have capacity to biotransform chlordiazepoxide to its active metabolite, N-desmethyl chlordiazepoxide. The transformation of chlordiazepoxide was inspected in the present study using fermentation method and HPLC analysis. Six fungi were incubated on rotary shaker for 72 hrs as per protocol and analysed their extracts by HPLC method. Among six, the sample of Aspergillus ochreus has shown an extra peak in HPLC compared to their respective controls (Figure 2). Hence, this was designated that it has capacity to biotransform chlordiazepoxide to its metabolite, CDM, when samples of other organisms were correlated with their controls, identical peaks were found in samples and controls indicated no metabolite was formed by other organisms. (Table 1)
Metabolite-CDM
The sample of Aspergillus ochreus has shown an extra peak at retention time of 6.9 min. when compared with its controls as given in Table 1 and in Figure 2. The extra peak indicated the formation of chlordiazepoxide metabolite, CDM by Aspergillus ochreus. Mass spectrum of chlordiazepoxide metabolite [CDM] formed by Aspergillus ochreus [Figure 4] has shown 15Da lower molecular weight than that of chlordiazepoxide [Figure 3] which suggested the loss of methyl group from parent drug structure. The structure of metabolite CDM was also supported by the mass fragments at m/z 250 and 269 [Figure 5]. 1H NMR spectrum of CDM has shown peak at δ 8.59 represented the presence of NH2 group [Figure 7]. The peak at 0.79 which is a representative of protons in methyl group is present only in proton NMR spectrum of Chlordiazepoxide and not identified in the proton NMR spectrum of CDM. Whereas the formation of free amino group in CDM suggested loss of methyl group at amino position indicated N-desmethylation. So, the metabolite CDM was confirmed as N-desmethyl chlordiazepoxide.
1H NMR proton assignment of chlordiazepoxide and metabolite was also supported the structure of metabolite [Table 2]. The metabolite formed was N- desmethyl chlordiazepoxide, an active metabolite of chlordiazepoxide. Hence, the formation of an active metabolite of chlordiazepoxide by Aspergillus ochreus was observed from above results.
Chlordiazepoxide is extensively metabolised to an active metabolite ‘N-desmethyl chlordiazepoxide’ by in-vitro liver microsomes in human.36 and other mammals37,38 similar to metabolite acquired in case of Aspergillus ochreus in the present study.
Metabolism induction and inhibition studies
CYP 3A4 enzyme is involved in mammalian metabolism of chlordiazepoxide to form N-desmethyl chlordiazepoxide. Hence, whether the enzyme like CYP3A4 is involved in fungal metabolism of chlordiazepoxide or not was examined by induction and inhibition studies using CYP 3A4 inducer (carbamazepine) and inhibitor (fluoxetine). The enzymes involved in biotransformation of chlordiazepoxide by Aspergillus ochreus to form N-desmethyl chlordiazepoxide was confirmed by induction and inhibition studies by calculating the percentage of metabolite formed during induction and inhibition studies.
Metabolite-CDM
In case of samples incubated with Aspergillus ochreus, the percentage metabolite (CDM) formation was increased in the presence of CYP 3A4 inducer and decreased in the presence of CYP 3A4 inhibitor [figure 8 and 9, Table 3]. It indicated that CYP 3A4 like enzyme might be involved in the fungal biotransformation of chlordiazepoxide by Aspergillus ochreus to form ‘CDM’, an active metabolite N-desmethyl chlordiazepoxide.
It is confirmed that, the involvement of CYP 3A4 like enzyme in fungal biotransformation of chlordiazepoxide similar to mammals.
Enzyme kinetic studies
The enzyme kinetic studies are vital to find out the affinity of fungal enzyme towards the substrate by estimating kinetic parameters like Km and Vmax.
Metabolite-CDM
The enzyme kinetic studies of fungal metabolism mediated by enzymes in Aspergillus ochreus to form CDM revealed that, as the incubation time was increased the percentage metabolite formation was also increased along with increase in the substrate concentration in the metabolism of chlordiazepoxide (Table 4). Velocity of metabolic reactions for six concentrations were obtained from slope values (Table 3) of lines found in graphs plotted between incubation time and percentage of metabolite (CDM) As per the results of study of effect of incubation time on percentage of metabolite (CDM) for six different concentrations of substrate, the percentage of metabolite, CDM formation was maximum for 72 hrs of incubation when compared with 24,36,48 and 60 hrs of incubation (Figure 10). The study of effect of substrate concentration on percentage of metabolite CDM formation revealed that, the maximum percentage of metabolite (CDM) was observed at 60 µg/ml concentration for 72 hrs of incubation. (Figure 11) The Michaelis–Menten kinetics was applied to biotransformation kinetics of CDM formation because, the velocity of reaction was increased with increase in the substrate concentrations from 10 µg/ml to 60 µg/ml. Km and Vmax values of metabolism reactions mediated by enzymes in Aspergillus ochreus to form CDM were 1.92 µg/ml and 0.13 µg/ml/hr respectively (Figure 12 and Table 5). These values stated that metabolism reaction which formed CDM proceeded with the maximum velocity (Vmax) of 0.13 µg/ml/hr, and half maximum velocity of reaction (Km) was found at the concentration of 1.92 µg/ml.
Km is the substrate concentration at which half the active sites of enzyme are filled by substrate. An enzyme with a high Km has a low affinity for its substrate, and a greater concentration of substrate is required to achieve Vmax and vice versa.39
Km value for formation of metabolite, CDM was less (1.92 µg/ml) so, the enzymes present in Aspergillus ochreus responsible for formation of CDM has shown more affinity towards chlordiazepoxide and low concentrations of chlordiazepoxide is enough to saturate the enzyme and to reach maximum rate of reaction to form metabolite CDM.
The maximum rate, Vmax, is the designate for number of substrate molecules transformed into metabolite by an enzyme molecule per unit time when the enzyme is fully saturated with substrate. It was found that rate of metabolic reaction was increased with increase in the substrate concentration, but when the rate of reaction reached to Vmax, further increase in the substrate concentration will not further increase in percentage of metabolite (CDM) formation. It indicated that the enzyme present in this fungus responsible for metabolism of Chlordiazepoxide was fully saturated with given substrate concentration of 60 µg/ml at Vmax.
It can be concluded that, Aspergillus ochreus has the potential to transform chlordiazepoxide to its active metabolite due to the presence of enzymes crucial for biotransformation of chlordiazepoxide. Formation of the active metabolite was similar to mammals. Induction and inhibition studies also confirmed that, CYP 3A4 like enzyme might be involved in the biotransformation of chlordiazepoxide by Aspergillus ochreus. Thus, the present study revealed that, this fungus can be used as a model for CYP3A4 enzyme to study pharmacological and toxicological properties and the biotransformation of other similar drugs. Enzyme kinetic studies revealed that, metabolism mediated enzymes present in Aspergillus ochreus possessed low Km value and has more affinity towards chlordiazepoxide, which in turn indicated that high amount of metabolite can be produced with less quantity of chlordiazepoxide.
ACKNOWLEDGMENTS
The authors would like to thank Department of Science and Technology, Government of India for Facilitating HPLC (DST-CURIE) and Milliporewater system (DST-FIST). The authors are also thankful to Sri Padmavathi Mahila Visva Vidyalayam for providing all the facilities to carry out this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MK compiled information from the literature, designed the figures and tables and drafted the manuscript. MV supervised and reviewed the manuscript. Both authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Raymond G. Booth. Psycho therapeutic agents:Antipsychotic and anti anxiolytic agents, In Williams, David H, Foye, William O, Lemke, Thomas L. 7th ed, Foye’s principles of medicinal chemistry. Hagerstown, MD: Lippincott Williams & Wilkins. 2012:623.
- Tong WY, Dong X. Microbial biotransformation: recent developments on steroid drugs. Recent Pat Biotechnol. 2009;3(2):141-153.
Crossref - Yang W, Ye M, Huang F, He W, Guo D. Biocatalysis of cycloastragenol by filamentous fungi to produce unexpected triterpenes. Adv Synth Catal. 2012;354(2-3):527-539.
Crossref - Sedlaczek L. Biotransformations of steroids. Crit Rev Biotechno. 1988;7(3):187-236.
Crossref - Bhatti HN, Khera RA. Biological transformations of steroidal compounds: a review. Steroids. 2012;77(12):1267-90.
Crossref - Perkins C, Siddiqui S, Puri M, Demain AL. Biotechnological applications of microbial bioconversions. Crit Rev Biotechnol. 2016;36(6):1050-1065.
- Kapoor A. Pharmacokinetics and Pharmacodynamics. In: Tampi R, Tampi D, Young J, Hoq R, Resnick K. (eds) Absolute Geriatric Psychiatry Review. Springer, Cham. 2021.
Crossref - Coelho, Maria A, Ribeiro, Bernardo D. White Biotechnology for Sustainable Chemistry,Green Chemistry Series, The Royal Society of Chemistry. 2016:36-51.
Crossref - Gauthier L, Bonnin-Verdal MN, Marchegay G, et al. Fungal biotransformation of chlorogenic and caffeic acids by Fusarium graminearum: New insights in the contribution of phenolic acids to resistance to deoxynivalenol accumulation in cereals. Int J Food Microbiol. 2016;221:61-68.
Crossref - Olicon-Hernandez DR, Gonzalez-Lopez J, Aranda E. Overview on the Biochemical Potential of Filamentous Fungi to Degrade Pharmaceutical Compounds. Front Microbiol. 2017;8:1792.
Crossref - Ricardo B. Biotransformation. Marine Pollution. 2018:205-214.
Crossref - Zhang Z, Tang W. Drug metabolism in drug discovery and development. Acta Pharm Sin B. 2018;8(5):721-732.
Crossref - Jurgens FM, Behrens M, Humpf H-U, Robledo SM, Schmidt TJ. In Vitro Metabolism of Helenalin Acetate and 11α13-Dihydrohelenalin Acetate: Natural Sesquiterpene Lactones from Arnica. Metabolites. 2022;12(1):88.
Crossref - Tomonaga S, Kawase T, Tsukahara T, Ohta Y, Shiraishi J-I. Metabolism of Imidazole Dipeptides, Taurine, Branched-Chain Amino Acids, and Polyamines of the Breast Muscle Are Affected by Post-Hatch Development in Chickens. Metabolites. 2022;12(1):86.
Crossref - Khojasteh SC, Argikar UA, Cho S, et al. Biotransformation novel advances – 2021 year in review. Drug Metab Rev. 2022;54(3):207-245.
Crossref - Chong S, Agarwal S, Agarwal S, et al. The nonclinical disposition and PK/PD properties of GalNAc-conjugated siRNA are highly predictable and build confidence in translation to man. Drug Metab Dispos. 2021;50(6):781-797.
Crossref - Agarwal S, Simon AR, Goel V, et al. Pharmacokinetics and pharmacodynamics of the small interfering ribonucleic acid, givosiran, in patients with acute hepatic porphyria. Clin Pharmacol Ther. 2020;108(1):63-72.
Crossref - Ma Y, Sun P, Zhao Y, et al. A Microbial Transformation Model for Simulating Mammal Metabolism of Artemisinin. Molecules. 2019;16;24(2):315.
Crossref - Wongjiratthiti A, Yottakot S. Utilisation of local crops as alternative media for fungal growth. Pertanika J Trop Agric Sci. 2017;40(2):295-304.
- Jain A, Jain R, Jain S. Sub-culturing of Bacteria, Fungi and Actinomycetes. Basic Techniques in Biochemistry, Microbiology and Molecular Biology, Springer 2020:101-103.
Crossref - Brandt P, Garbe E, Vylkova S. Catch the wave: Metabolomic analyses in human pathogenic fungi. PLoS Pathog. 2020;16(8):e1008757.
Crossref - Gottschalk LA, Kaplan SA. Chlordiazepoxide plasma levels and clinical responses. Compr Psychiatry. 1972;13(6):519-527.
Crossref - Kavitha K, Vidyavathi M, Asha S, Hima Bindu TVL. Microbial Metabolism and Inhibition Studies of Phenobarbital. J Pharm Res. 2012;11(1):62-68.
Crossref - Yuh L, Ping L, Cuyue T, et al. Substrate Inhibition Kinetics for Cytochrome P450-Catalyzed Reactions Part 1. Drug Metab and Dispos. 2017;29(4):368-374.
- Cusinato DAC, Filgueira GCO, Rocha A, Cintra MACT, Lanchote VL, Coelho EB. LC-MS/MS analysis of the plasma concentrations of a cocktail of 5 cytochrome P450 and P-glycoprotein probe substrates and their metabolites using subtherapeutic doses. J Pharm Biomed Anal. 2019;164:430-441.
Crossref - Sujana K, Hamuthal MZV, Murthy VSN, Shravani N. A Novel Validated Analytical Method Development for the Binary Mixture of Mebeverine and Chlordiazepoxide in Pharmaceutical Formulation and its Application to Stress Studies. Pharm Anal Acta. 2015;6(1):324.
Crossref - Patel SK, Patel NJ. Simultaneous RP-HPLC Estimation of Trifluoperazine Hydrochloride and Chlordiazepoxide in Tablet Dosage Forms. Indian J Pharm Sci. 2009;71(5):545-547.
Crossref - Ashour S, Kattan N. Simultaneous Determination of Clidinium Bromide and Chlordiazepoxide in Combined Dosage Forms by High-Performance Liquid Chromatography. J Pharm (Cairo). 2013;2013:417682.
Crossref - Strojny N, Puglisi CV, de Silva JAF. Determination of Chlordiazepoxide and Its Metabolites in Plasma by High Pressure Liquid Chromatography. Analytical Letters. 1978;11(2):135-160.
Crossref - MacDonald A, Michaelis AF, Senkowski BZ. Chlordiazepoxide. Analytical Profiles of Drug Substances. 1972;1:15-37.
Crossref - Roskar R, Lusin TT. Analytical methods for quantification of drug metabolites in biological samples. Chromatography-The Most Versatile Method of Chemical Analysis. 2012:79-126.
Crossref - Segel IH. Enzyme Kinetics: Behaviour and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. (ed.Wiley & Sons Inc) New York. 1975;8(1):159-162.
- Magang S, Lin Y, Lu P, et al. Enzyme Kinetics of Cytochrome P450-Mediated Reactions. Curr Drug Metab. 2000;2(1):17-36.
Crossref - Palva ES, Linnoila M. Effect of active metabolites of chlordiazepoxide and diazepam, alone or in combination with alcohol, on psychomotor skills related to driving. Eur J Clin Pharmacol. 1978;13(5):345-350.
Crossref - Ashton CH. Benzodiazepine equivalency table. 2007. http://www.esculape.com /medicament/Benzodiazepines-equivalences.pdf
- Koechlin BA, Schwartz MA, Krol G, Oberhansli W. The metabolic fate of C14-labeled Chlordiazepoxide in man, in the dog, and in the rat. J Pharmacol Exp Ther. 1965;148(3):399-411.
- Datta PR, Nelson MJ. Enhanced metabolism of methyprylon, meprobamate, and chlordiazepoxide hydrochloride after chronic feeding of a low dietary level of DDT to male and female rats. Toxicol Appl Pharmacol. 1968;13(3):346.
Crossref - Wang LL, Ren XX, He Y, et al. Pharmacokinetics of Diazepam and Its Metabolites in Urine of Chinese Participants. Drugs R D. 2022;22(1):43-50.
Crossref - Reuveni S, Urbakh M, Klafter J. Role of substrate unbinding in Michaelis-Menten enzymatic reactions. Proc Natl Acad Sci USA. 2014;111(12):4391-4396
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.