ISSN: 0973-7510
E-ISSN: 2581-690X
Biopolymer of chitin and chitosan were extracted from the two Pleurotus spp. of P.floridanus and P.djamor. After the optimization, genomic DNA was isolated, and by using ITS 1 and ITS 4 primers molecular identification was carried out and the nucleotides were submitted to NCBI. The biopolymer have a great commercial importance now a days because of it high efficiency in all the field. The present study was focused on to extract the chitin and chitosan from the Pleurotus spp. and these polymer was subjected to analysis the total antioxidant activity, antimicrobial activity against food borne pathogens of E.coli, B. cereus, S.aureus, A.niger and A.flavus. Cytotoxicity study against HeLa Cell Line was also performed to study the efficacy of the extracted polymer.
Pleurotus spp, chitin, chitosan, total antioxidant, antimicrobial activity, Cytotoxicity
Chitin is similar to cellulose with the chemical structure of [poly-b-(1 ® 4) – N-acetyl-D-glucosamine], chitosan is one hydroxyl group on each monomer has substituted with an acetyl amine group.1 These polysaccharide is chiefly present in the exoskeleton of crustaceans (shells of crab, shrimp, prawn, etc) and contain a percentage of inorganic material CaCO3, while production which is releasing CO2 to the environment. The present pertain about the global warming which is not an advisable or environmental friendly process.2
Several method was already done to extract the chitin and chitosan from the crustaceans, despite these have thousands of applications and use in the field of medical and industrially. The importance and the needs of these polymers also facing the problem of seasonal variations and it is non-vegetarian source. To put back these problems scientist find an alternative source to extract these polymer from some plants and fungi or from mushroom.
Chitin is widely distributed in fungi where it is component of the cell wall and structural membrane of mycelia, stalks, and spores. Chitin in fungi possess principally the same structure of chitin occurring in other organism.3However that all fungi contain chitin and the polymer may be absent in one specious that is closely related to the other variation. In the amount of chitin may depend on physiological parameters, environmental and fermented conditions, processing and in the culturing of the fungi.
The importance and use of chitin and chitosan is increasing day by day is because of its antioxidant activity, 4 biocompatibility of which does not have an toxic or injurious effects on biological system, 5 biodegradability – which is being able to broken down by natural process that is by bacteria, fungi, and other simple microorganism,6 antimicrobial activity, 7 cosmetics, medicinal applications, food, flavour or micro encapsulater, flocculatants for liquid wastes,8 enzyme immobilizer, production of contact lens and production of eye bandage.9
The advantage or benefits of using fungi is the easy handling, harvesting and controlling to produce high quality chitin and chitosan. Guha et al10 reported that the different type of fermentation technologies are helping to replace these problems by concerning the conventional method to produce these polymers from fungi.
The present study was aimed on to extract the chitin and chitosan from the Pleurotus spp. The Molecular identification of Pleurotus spp. was done followed by the extraction of chitin and chitosan. After characterisation total antioxidant activity, antimicrobial activity against food borne pathogens and cytotoxicity study against HeLa Cell Line were performed.
Source of inoculums
The two Pleurotus spp. Strain were collected from Tamilnadu agricultural university, Coimbatore, Tamilnadu, India and brought to the laboratory in an air tight container. The strain was subculture to PDA plate (39gm in 1000ml, Himedia, Mumbai, India) and incubated at 300C for 10-12 days. After mycelial growth mycelia was separated and used for further studies.
Molecular identification
DNA isolation
DNA isolation was done by the modified protocol of Kumar et al11 the growed mycelia was crushed with CTAB buffer and incubated at 650C for 45 minutes. After incubation the supernatant was collected by centrifugation. Chloroform and isoamyl alcohol was added in the ratio of 24:1 and allowed for centrifugation to separate the aqueous layer. To the collected aqueous layer 2 volume of isopropanol was added and pellet was collected by centrifugation. Pellet was washed with absolute ethanol and 70% ethanol to remove the impurities. After air drying the pellet was dissolved by adding 1X TE buffer and the isolated DNA was confirmed by 0.8% agarose gel.
PCR and Sequencing
The isolated DNA was used for molecular identification using AB Applied Biosystem veriti 96well Thermal cycle PCR detection system. For the amplification, universal fungal primer of internal transcribed spacer 1 (ITS1:5′-TCCGTAGGTGAACCTGCGG-3′) and internal transcribed spacer 4 (ITS4:5′-TCCTCCGCTTATTGATATGC-3′) were used. To the 2µl of isolated DNA1µl of each ITS primers,10µl of the master mix (Sigma), and 8µl of distilled water was added to prepare the PCR reaction mixture. PCR was done, initial denaturation at 950C for 2minutes, denaturation 950C for 1minutes, annealing at 550 C for 1 minutes, and extension at 720C for 1minutes and final extension for 5 minutes at 720C the total number of cycle was 30. The amplified product was analysed by 1.5% agarose gel. The PCR product was purified using Quiagen kit as per the manufacture instruction and sent to Chromous Biotech Pvt. Ltd Bangalore for sequencing. The sequence result was analysed by BLAST, and submitted to NCBI for accession number and the phylogenetic tree was constructed using MEGA-7 software.
Production and extraction of chitin and chitosan in fermentation medium
The production of chitin and chitosan was done from the mycelia mat of both blended Pleurotus spp. which was grown in the MGYP (Meat extract-2%, Glucose –1.5%, Yeast –0.3%, Peptone –0.5%) medium. After 10-12 days of incubation described by Jesteena et al12. The mycelia mat was separated aseptically and the polymer was extracted from both the Pleurotus spp. by the treatment method of alkaline, acid and deproteinasation to get the chitin. Chitosan was extracted from the chitin by deacetylation to precipitate the polymer Sandra et al13. The final product was washed with ethanol and allowed for drying, after drying the sample was used for characterisation and further applications.
Characterisation of extracted chitin and chitosan
UV-VIS study
The extracted chitin and chitosan sample was further confirmed by UV-Visible (Labtronics Microprocessor) study in the nanometer of 200-500nm, after setting the baseline with the scanning interval of 2nm.
FTIR -Fourier Transform Infrared Spectroscopy
To analyse the functional group which is present in the chitin and chitosan sample, the scan was done in the spectral region of 500cm-1 to 4000cm-1 with the resolution of 4cm-1 using Shimadzu FTIR instrument. The dried sample was prepared by kBr pellet and stabilized before undergoing the spectrum analysis.
Total antioxidant activity
Total antioxidant activity of the extracted sample was analysed by phospho molybdenum method described by Prieto et al14 with slight modification. The 1ml of the extracted chitin and chitosan (0.1 g/ml) was mixed with equal amount of reaction mixture (0.6M H2SO4, 28mM sodium phosphate and 4mM ammonium molybdate). The addition of reaction mixture, sample was incubated at 450C for 90minutes in water bath. After incubation the sample was allowed to cool in room temperature and the OD measurement was measured at 695nm using UV-Visible spectrophotometer (ELICO SL 159). Blank was prepared without the addition of sample and the mg/g of total antioxidant was calculated by using ascorbic acid as a standard.
Antimicrobial activity
The antimicrobial activity was done by well diffusion method against food borne pathogens of, E.coli, S.aureus and B.cereus for antibacterial and antifungal activity against A.niger, and A. flavus, by the previous method of Jesteena et al.12Cefotaxime (CTX 30) disc was used as a positive control for bacteria, fluconazole (30mg/ml) was used for fungi. After specific incubation period the zone of inhibition was measured.
Cytotoxicity study
Cytotoxicity study was carried out using MTT(3-(4,5-Dimethylthiazol-2-yl)-2,5diphenyletrazolium bromidefor) assay method by Grushree et al.15 HeLa Cell Line in DMEM medium was allowed to grow in the CO2 incubator by 5% CO2, 80% humidity and at 370C. After 48hrs of incubation the live cells was confirmed by inverted microscope (Unicon) and the cells was transferred to microplate reader. The sample in the different concentration of 10,20,30,40,50µl was taken along with 100µl of the Cell Line, control (only Cell Line) also added and incubated in the CO2 incubator for 24 hrs. After incubation the cells was washed with DMSO and trypsin. 20µl of MTT dye was added and using ELISA (Robonik-Readwell Touch Elisa Plate Analyser) reader at 570nm the OD was taken after 24hrs. The percentage of cell death was calculated using the following formula;
% of cell death = [Control OD –Sample OD / Control OD] × 100
The optimum day was confirmed by the report of other research article and also the previous work done Jesteena et al.12 Sandra et al13 reported that the maximum biomass production was obtained after 8th day of incubation 21.87±2.2g/L-1. The same was incubated upto 14th days but the substrate decreased during the day of culture until a minimum of 5.57±0.85g/L-1. Maghsoodi et al16 observed the maximum production of chitosan was in the day of 12th and the production was started at the day of 4th. In the present study after 10-12th day of incubation, it shown good mycelial growth and this was used for molecular identification and also the extraction of chitin and chitosan.
Molecular identification
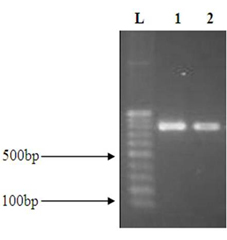
The DNA of the both Pleurotus spp. was done separately and was documented on 0.8% agarose gel. The result was given in figure 1.
Fig. 2. PCR result of the two white rot fungi Lane 1:100Kbs Ladder. Lane 2: P.floridanus Lane 3: P.djamor
The molecular analysis of partial DNA gene sequencing shows 88 % of similarities with P.floridanus and 83% similarities for P.djamor and the NCBI accession numbers were MG324371.1 and MG328900.1
Sasidhara and Thirunalasundari17 done molecular identification of the mushroom using genomic DNA with the approximate molecular weight of 559 base pairs with ITS1 and ITS4 primers and submitted to the gene bank in the name of Agaricomycetes Spp. II India 01, these wild mushroom shows the homology score of 88 with Perenniporia Spp. Alvardo et al18 done the DNA extraction from the mycelium to get higher quantity of the genetic material and they used ITS1 and ITS2 for their PCR cycling.
Menolli et al19 reported the molecular identification of P. ostreatus with 90% support with other sequence from the NCBI gene bank P.ostreatus and P.pulmonarius. Chandra et al20 constitute that the similarities of 27% between P.floridanus and P.sajorcaju. This is may be because of different cultivated area or based upon the growth condition.
Characterisation
UV-Visible analysis
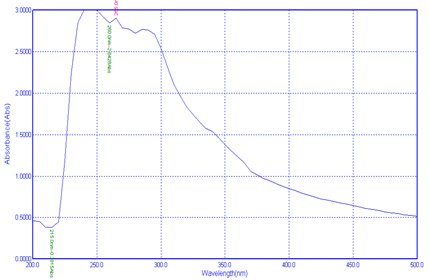
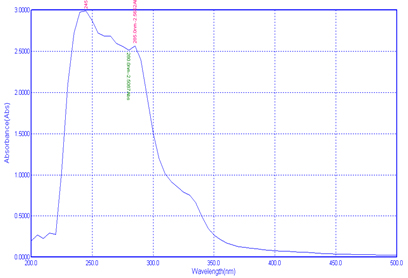
UV-Visible spectroscopy analysis is helping to identify the type of absorption from the ground stage to excited stage. In the present study the polymer was scanned at 200-500nm was used to analysis the presence of extracted chitin and chitosan from the Pleurotus spp. The study was showing three Plasmon peak for chitin in the nm of 265.0nm, 260.0nm and 215.0nm (Fig 5).For chitosan 285.0nm, 280.0nm and 245.0nm(Fig 6).
The earlier report by Krishnaveni and Ragunathan21 observed the Plasmon peak at 390nm for their synthesised nanocomposites from the chitin chitosan. Negrea et al22 got minimum absorption of 212.6nm for chitin, and for Silva et al23 got peak on 202nm.
FTIR
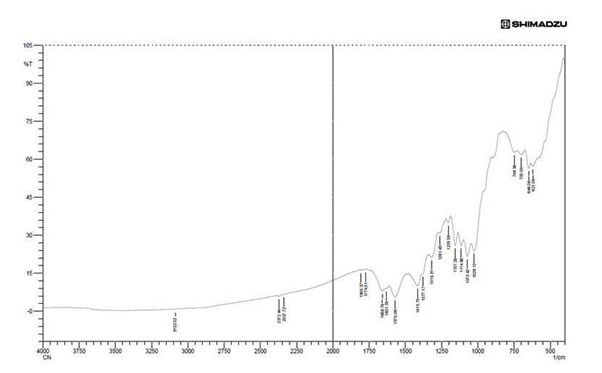
In the analytical part infrared spectrum field was 4000cm-1-500cm-1 and the resulting band was larger macromolecular character because of the inter molecular binding of hydrogen. Tasar et al24 in their structural group identification FTIR was carried out from 4000cm-1 – 550cm-1 and they are finalised for industrial purpose with large production of high quality chitosan from low cost method.
FTIR result of chitin (Fig.7) showed the presence amide III in 952.84cm-1 to 1261.45cm-1amide III acetyl group. From the result of Prabhu and Natarajan25 the band of 1261.45 is an acetyl ester band, 1570.06cm-1 is an N-acetyl ester band and usually an high amide III absorption. 1631.78cm-1 to 1658.78cm-1 is an amide C=O stretch, 1774.51 is an ketone C=O stretch absorption is an strongest IR absorption and these are use full in the structure determination of chitin.3102.01cm-1 is an amino peak alpha chitin and which is also an stretching of alkene C-H which correspond to the vibrational stretching of hydroxyl group of our study.
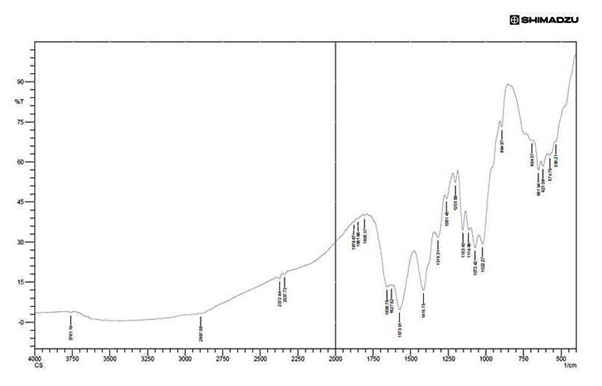
Fig.8 shows the chitosan sample showed bands at 3761.91cm-1 due to the stretching vibration of OH group 2897.08cm-1 is the vibration of CH bond (aldehyde) Silva et al.26 Stretching at 1658.78cm-1is the vibration of alkene group, the stretching from 1072.42 cm-1 to 1026.13 cm-1 has been assigned to vibration of CO group Xu et al.27 The band located at 1153.43 cm-1 (C-O-C vibration) is an vibration of CO in the oxygen bride is the outcome of deacetylated chitosan Silva et al.26 The peak at 894.97 showing the presence of polysaccharide structure of chitosan and the band from 1072 cm-1 showing the binding of C-O-H, C-O-C, CH2CO Paluszkiewicz.28 Other band which is formed before 894.97 is showing the strong bending of C-H group, and in the particular study bands of 1415.75 cm-1 was the vibration of O-H group (alcohol), 2897.08 is the vibration of C-H stretching (aldehyde) and 2372.72 cm-1 is a strong O=C=O stretching.
Total antioxidant activity
Using phospho molybdenum method the total antioxidant activity of the sample was quantified for chitin and chitosan. Total antioxidant activity of the sample of chitin 101mg/g and for chitosan 138mg/g. According to Prieto et al14 the absorbance at 695nm showing phospho molybdenum complex reaction from MO (V1) reduction to MO (V) which is turned to blue green colour.
Xie et al29 reported that the antioxidant activity of the chitosan may be due to the reaction of residual free or free radical or the amino group to form constant macromolecule radicals or the group can form amino group by absorbing H2 ions from the mixture and then reacting with radical through can additional reaction.
Antimicrobial activity
Antibacterial activity of the two extracted polymer was showing good result against the food borne pathogens. For all the selected microorganisms, chitosan showing good result compared with chitin. The results were given in the Figure 9 and 10, Table 1.
Table (1):
Zone of inhibition in mm for antimicrobial activity of the extracted sample.
| Sample used | Food pathogens used | ||||
|---|---|---|---|---|---|
| Bacteria | Fungi | ||||
| B.cereus | E.coli | S.aureus | A.niger | A.flavus | |
| Chitin (cn) | 2 mm | 7 mm | 3 mm | 3 mm | 2 mm |
| Chitosan (cs) | 5 mm | 10 mm | 6 mm | 3 mm | 1 mm |
| Antibiotic | 1 mm | 12 mm | 2 mm | 4 mm | 3 mm |
Fig. 9. Antibacterial activity against B.cereus, E.coli and S.aureus from the, figure cn-chitin, cs-chitosan, D- antibiotic disc
Sudharshan et al30 investigated the antimicrobial activity of the various water soluble chitosan on dissimilar gram negative and gram positive bacteria and also against fungi which shown good result in a very short time. Research of Shaidi et al31 reported that this changes is because of the electro negatively charged site of bacterial surface causing some agglutination in the chitosan treated cells, were the interaction between the bacteria (especially E.coli and Salmonella Spp.) it is showing non- liberation of lipopolysaccharide in other lipid from the membrane analysing with electronmicroscopy showing which will cause exterior modification in the bacterial cells.
Fig. 10. Antifungal activity against A.niger and A.flavus from the fig-ure,cn-Chitin, cs-Chitosan, Ab- Antibiotic
Nadarajah and Cheng 32 reported that the chitosan have a good antibacterial activity against gram positive and gram negative bacteria. In their investigation 2-10mg/ml was used and 8mg/ml shows higher percentage of inhibition against gram negative bacteria of B.cereus, P.aeruginosa and Enterococcus aerogenes. The previous results of the antimicrobial activity of the chitin and chitosan extracted from the Pleurotus spp. Jesteena et al12 also showed better result against E.coli, S.aureus and B.subtilis. Antifungal study showed better result against A.niger, A.flavus and F.solani.
According to Ghaouth et al7 chitosan have the ability to conquer to inhibit the growth of fungi. It delayed the acceleration of the decay of stored fruits. Furthermore chitosan coated fruits inhibit the growth of P. digitatum, B. lecanidion, B. cinerea and P.italicum in citrus fruits Chien et al. 33
Cytotoxicity study
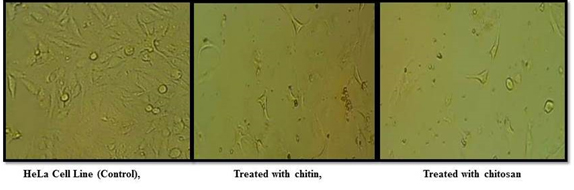
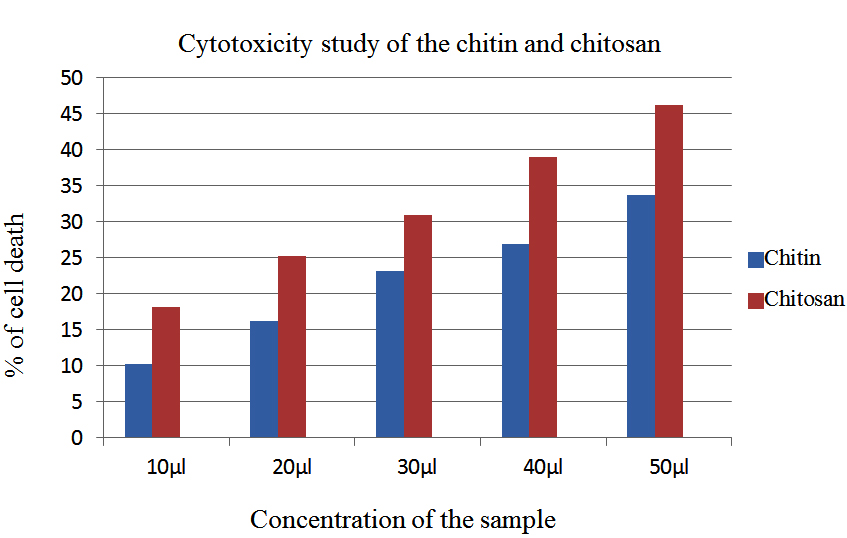
This research was done to identify the cytotoxicity in the aim of finding an alternative source to kill the cancer cells. The percentage of cell death was increasing with increasing concentration for both the sample. For chitin 10.25, 16.15, 23.15, 26.89, 33.75 % of cell death, and for chitosan 18.16, 25.58, 30.96, 39.02, 46.19% of cell death from the concentration of 10 µl to 50µl of the sample was occurred. Compared to chitin,chitosan showing better results against the Cell Line. Fig.11 denoting microscopic view of the control Cell Line, cell death by the addition of 50µl of chitin and 50 µl of chitosan sample.
Fig. 11. Control Cell Line and the Cell Line treated with chitin and chitosan sample – micro-scopic view
From the review of Yoshinori et al34 chitin and chitosan derivatives are campaigner to carry anticancer agent. The conjugated chitosan demonstrating good result in the life span of P388 leukemia bearing mice after the intraperitoneal injection. On the other hand the result of Leena et al35 stated that the chitosan concentration from 1000 µg/ml to 7.8µg/ml and they got 50% of cell death against A549 lung cancer Cell Line using MTT assay method.
The present research can be resolved effectively to produce the chitin and chitosan from the Pleurotus spp. Culturing in MGYP medium, the optimization was confirmed by using the formation of mycelia mat on the media. The two Pleurotus spp was subjected to molecular level identification to confirm the fungi and got 88% similarities to P.floridanus and other fungi shown 83% similarities to P.djamor. The mycelia mat after 12th day of incubation blended and used to extract the chitin and chitosan. The extracted chitin and chitosan were characterised by UV-Visible study and FTIR to finalise the functional group. So far limited paper only reviewed about the antioxidant activity and this work is unique to quantifying the antioxidant activity and anticancer study. Total antioxidant activity using phosphor molybdenum method for chitin 101mg/g and for chitosan138mg/g. Antimicrobial activity against the food borne pathogens of E.coli, B.cereus, S.aureus, A.niger and A.flavus displaying better result, cytotoxicity study also showing 10.25 to 46. 19 % of cell death against the HeLa Cell Line. Discussing with these applications, chitosan showing dependable results comparing with chitin for total antioxidant activity, antimicrobial activity and cytotoxicity study.
ACKNOWLEDGMENTS
The authors acknowledge the management and staff of Centre for Bioscience and Nanoscience Research, Coimbatore – 21, provided the well-equipped infrastructure for the success full completion of this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Ravikumar, M. N. V. A review of chitin and chitosan application. React Funct Polym. 2000; 46: 1-27.
- Roberts George. European chitin society newsletter. Truth or Myth? UK; 2006.
- Peter M G. chitin and chitosan in fungi. Biopolymers online. 2005;6: 123-157.
- Sun, T., Yao, Q., Zhou, D., Mao, F. Antioxidant activity of N-carboxymethylchitosan oligosaccharides. Bioorg Med Chem Let. 2008; 18: 5774-5776.
- Pradip, Kumar, Dutta,. Joydeep, Dutta., V S Tripathi. Chitin and chitosan: chemistry, properties and application. J Sci Ind Res. 2004;63: 20-31.
- Kean, T., Thanou, M. Biodegradation, biodistribution, and toxicity of chitosan. Adv Drug del Rev. 2010; 62: 3-11.
- Ghaouth,E. I., Smilanick, A. J. L., Wilson, C. L. Enhancement of performance of Candida saitoana by the addition of glycol chitosan for the control of postharvest decay of apple and citrus fruit. Post Harvest Bio Tec. 2000; 19: 103-110.
- Jeon, Y. J., S. K. Kim., Effect of antimicrobial activity by chitosan oligosaccharide N-conjugated with aspargine. J Micro Biotech. 2001; 11:281-286.
- Muzzarelli, R. A. A. Chitin. Pergon press. Oxford. UK. 1997.
- Guha, A. K., Chatterge, B. P., Chattergee, S., Adhy, M. Chitosan from Mucor rouxii: production and physio-chemical characterization. Proc Bio Chem. 2005; 40: 395-400.
- Kumar, M. S., G. Kaur., A. K. Sandhu. Genomic DNA isolation from Fungi, algae, plant, bacteria and human blood using CTAB method. Int J Sci Res. 2014; 3:617-618.
- Jesteena, Johney., Kannan, Eagappan., R. Ragunathan. Microbial extraction of chitin and chitosan from Pleurotus spp, its characterization and antimicrobial activity. Int J Cur Pharm Res. 2017; 9 (1): 88-93.
- Sandra, Patricia, Ospina, Alvarez., David, Alexander, Ramirez, Cadavid., Diana, Marcela, Escobar, Sierra., Claudia, Patricia, Ossa, Orozco., Diego, Fernando, Rojas, Vahos., Paola, Zapata, Ocambo., Lucia, Atehortua. Comparison of extraction methods of chitin from Ganoderma lucidum mushroom obtained in submerged culture. Biomed Res Int. 2014. 1- 7.
- Prieto, P., Dinedu, M., Aguilar, M. Spectrophotometric Quantification of antioxidant capacity through the formation of a phosphor molybdenum complex: Specific application to the determination of Vit E1. Analytical Biochemistry. An open-access and academic publisher. 1999; 269(2):337-341.
- Gurushree Soundarajan., N. G. Ramesh Babu., Jesteena, Johney., R. Ragunathan. Extraction of bioactive compounds from Rosmarinus officinalis L. and its anticancer activity against HeLa Cell Line. Int J Sci Res.2017; 6 (8): 165-168.
- Maghsoodi, V., J. Razavi., Yaghmaei. Solid-state fermentation for production of chitosan by Aspergillus niger. Int J Eng Tran B Appl. 2009;22(1): 1-6.
- Sasidhara, Rajaratnam., Thirunalasundari, Thiagarajan. Molecular characterization of wild mushroom. Euro J Exp Biol. 2012; 2(2):369-373.
- Alvardo – Marchena, L., R. Chacon-Cerdas., C. Alvardo – Ulloa., A. Schmidt – Duran., D. Flores-Mora. Identification of Phytophthora spp. As a Causal agent of stem rot and dieback in fig trees of Costa Rica. APRN J Agri Bio Sci. 2017;12(10): 307-314.
- Nelson, Menolli, Junior., Tatiane, Asai., Marina, Capelari., Luzzia, Doretto, Paccola, Mierelles. Morphological and molecular identification of four Brazilian commercial isolates of Pleurotus spp. And cultivation on corncob. Brazilian J Bio Tec.2010;53 (2): 397-408.
- Chandra, S., Ghosh, K., Acharya, K., Comparative study on the Indian cultivated Pleurotus spp. by RAPD finger printing. J Nat Sci. 2010; 8: 90-93.
- Krishnaveni, B., R. Ragunathan. Extraction and characterization of chitin and chitosan from Aspergillus terrus spp. Synthesis of their bionanocomposites and study of their productive applications. J Chem Pharm Res.2015; 7(2): 115-132.
- Negrea, P. A., Caunii., Sarac, I., Butnariu, M., The study of infrared spectrum of chitin and chitosan extract as potential source of biomass. Dij J Nano Bio Str. 2015; 10 (4): 1129-1138.
- Da, Silva, R. M. P., Mano, J. F., Resis, R.L. Straight forward determination of the degree of N-acetylation of chitosan by means of first derivative UV spectrophotometry. Mac Chem Phy. 2008; 10 (4): 1463-1473.
- Tasar, O. C. S., Erdal, M. Taskin. Chitosan production by psychro tolerant Rhizopus oryzae in non sterile open fermentation conditions. Int J Bio Macromol. 2016; 89: 428-433.
- Prabhu, K., Natarajan, E., Isolation and FTIR spectroscopy characterization of chitin from local sources. Adv Appl Sci Res. 2012; 3(2): 1870-1875.
- Suedina, M. L. Silva., Carla, R. C. Braga., Marcus, V. L. Fook., Claudia, M. O. Raposo., Laura, Carvalho., Eduardo, L. Canedo. Application of infrared spectroscopy to analysis of chitosan/clay nanocomposites, Infrared spectroscopy-materials science. Eng Tec. 2012; 43-62.
- Xu, Y., Kim, K., Hanna, M. A., Nag, D. Chitosan starch composite film: preparation and characterization. Ind Crops Prod. 2005;21: 185-192.
- Paluszkiewicz, C., Stodolak, E., Hasik, M., Blazewicz, M. FTIR study of montmorillonite-chitosan nanocomposite materials. Spec Acta Part A. 2011;79: 784-788.
- Xie, W., Xu, P., Liu, Q., Antioxidant activity of water –soluble chitosan derivatives. Bioorg Med Chem Let. 2001;11: 1699-1701.
- Sudharshan, N. R., Hoover, D. G., Knorr, D. Antibacterial action of chitosan. Food Biotec. 1992; 6: 257-272.
- Shahidi, F., Arachchi, J. K. V., Jeon, K. J. Food application of chitin and chitosan. Trends food Sci Tec. 1999; 10: 37-51.
- Nadarajah, T. Y. W., Cheng. Isolation and characterization of fungal chitosan from Malaysian isolates. J Pure Appl Micro. 2009; 3(2): 1-8.
- Chien, P., J, Sheu, F., Lin, H. R. Coating citrus fruit with low molecular weight chitosan increases postharvest quality and shelf life. Food Chem. 2007;100: 1160-1164.
- Yohinori, Kato., Hiraku, Onishi., Yoshiharu, Machida. Contribution of chitosan and its derivatives to cancer chemotherapy. In Vivo. 2005;19: 301-310.
- Leena, Regi, Saleth., Shanthinee, Ravichandren., Seethalakshmilllanchezian, Ramasamy, Sivasamy. Anti-proliferative effect of Prawn Shells chitosan on lung cancer (A549) Cell Line. Int J chem Tech Res. 2014; 6 (9): 4102-4107.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.