ISSN: 0973-7510
E-ISSN: 2581-690X
Escherichia coli DH10B has 1.1 kb ybdK gene which is responsible for encoding YbdK enzyme that possess a Gamma glutamyl cysteine synthetase activity. ybdK gene was ligated downstream of a constitutive derepressed lac promoter of a low copy number plasmid vector pBBR1MCS-2, giving rise to a recombinant plasmid pPAT. Sinorhizobium fredii NGR234 transformed with pPAT showed an augmented production of glutathione which in turn increased the production of cadmium sulphide nanoparticles to some extent. Also, a heterologous expression of YbdK in Sinorhizobium fredii NGR234 improved the oxidation status of bacterial cells which is confirmed by fluorescence microscopy images and fluorometry. Genetically modified (GM) cells stained by DCFDA showed a significant decrease in fluorescence compared to wild type (WT) cells. Physical and chemical properties of the nanoparticles produced by the pPAT transformed Sinorhizobium fredii NGR234 differed significantly compared to wild type (WT) Sinorhizobium fredii NGR234. Comparative analysis of the nanoparticles by FTIR and SEM analysis revealed the functional groups attached to nanoparticles and average nanoparticle size respectively. Nanoparticles synthesized by genetically modified (GM) bacteria were about 3 times smaller in size compared to those produced by wild type (WT) rhizobium. FTIR analysis revealed an augmented presence of peptide with the nanoparticles produced by GM bacteria compared to those produced by the WT bacteria. XRD data revealed that biosynthesized CdS nanoparticles are face centered crystalline particles which was confirmed by comparing the peaks to standard JCPDS data (JCPDS card no. 10-454).
Sinorhizobium fredii NGR234, YbdK, Glutathione, Heavy metals, Cadmium sulphide, Nano particles, Green synthesis
Heavy metal pollution is one of the major environmental issues faced by the world. This problem is predominantly due to uncontrolled anthropogenic activities which has reached an alarming levels and it requires an immediate and drastic efforts to mitigate. Metals whose density is greater than 5g/cm3 are considered as heavy metals.1 Heavy metals can cause range of biological deformities and instabilities at molecular level including DNA damage, cell membrane damage and protein misfolding which can cause serious problems within the biological system by hampering the normal function of macromolecules.1 Research around the world is focused on developing new technologies and methods to tackle the problem of heavy metal pollution by green methods which can be broadly classified as Bioremediation.2 Microorganisms like fungi, bacteria and viruses plays a major role in bioremediation of heavy metals from the soil as well as water.2 Bacteria possess varieties of mechanisms such as extracellular barrier, efflux pump, intracellular /extracellular sequestration and reduction of metal ions to resist the presence of heavy metal in their surroundings.3 Many genus of soil bacteria and Plant growth promoting bacteria (PGPB) such as Mesorhizobium, Sinorhizobium and Bradyrhizobium have shown their ability to tolerate the presence of heavy metals like Cd, Co, Fe, Ni, Zn and Cu4 in their environment.
The remarkable ability of bacteria to reduce the metal ions thus converting them into less toxic forms is the best candidate for metal nanoparticle synthesis.5 Wild type as well as genetically modified bacteria have a potential to produce varieties of metal nanoparticles in in vitro conditions.5,6 Also Escherichia coli (MTCC10312) showed extracellular7 production of cadmium sulphide quantum dots. And a cyanobacteria Oscillatoria limnetica has shown its ability to produce silver nanoparticles.8 Not only wild type but genetically modified bacteria could be a good tool to manufacture nanoparticles in in vitro conditions. It was reported that genetically modified Escherichia coli over expressing glutathione synthetase showed augmented biosynthesis of cadmium sulphide nanoparticles.9 Also Escherichia coli expressing Candida albicans Metallothionine gene and Rhizobium tropici Phytochelatin synthetase gene showed enhanced production of silver nanoparticles10 and selenium nanoparticles11 respectively. Biogenic nanomaterial has unique physical and chemical properties and they are used in varieties of industries such as electronic, chemical, photonics, energy and medical.12 It is clear that microbe based nanoparticles production could be cheaper and environment friendly. Also, their unique properties allows its traditional as well as novel application in a wide spectrum of Industries.13 Genetically modified Escherichia coli strain coexpressing Metallothionine and Phytochelatin synthetase has been reported to produce 33 inorganic nanoparticles (20 single element and 13 multi-element ),14 which is way more than capacity of a synthetic reaction to produce that many inorganic nanoparticles in a single reaction. Additionally, the ease to genetically manipulate the microbes makes it a best candidate for production of nanoparticles in a way which consumes very less amount of energy thus contributing less in carbon foot print addition to our environment. Phytochelatin as well as Metallothionine have been extensively expressed/coexpressed/overexpressed for biosynthesis of nanoparticles for example PCS and ghs1 genes of Schizosaccharomyces pombe overexpressed in E.coli for CdS nanoparticles synthesis.15 Also an enhanced production of glutathione by Escherichia coli showed intracellular production of CdTe quantum dots.16 The probable mechanism of thiol containing proteins, amino acids and short peptides is to form metal thiolate conjugate with the heavy metals, which can be explained by nRSH + Mn+ ↔ (RS)n-M + nH+ equation.17
Glutathione synthesis in prokaryotes is governed by two enzymes, Gamma glutamyl cysteine synthetase (y-GCS) that catalyzes the ligation of glutamate and cysteine to form gamma glutamyl cysteine while the second enzyme glutathione synthetase catalyzes the ligation of gamma glutamyl cysteine and glycine to form a unique tripeptide glutathione.9 Glutathione has numerous functions such as metal chelation and alleviation of ROS inside the cells. Gamma glutamyl cysteine synthetase is a rate limiting enzyme of glutathione synthesis pathway, which binds to glutathione and controls its production by feedback mechanism whenever necessary. Escherichia coli DH10B YbdK, a carboxylate amine ligase also exhibits gamma glutamyl cysteine synthetase activity. It is a smaller protein with only 372 amino acids which possesses a different overall fold. Tertiary protein structure of YbdK is more similar to Glutamine synthase than bacterial y-GCS.18 So this study hypothesizes the use of structural dissimilarity of YbdK to hinder the feedback mechanism of glutathione and use a Sinorhizobium fredii NGR234 as a chassis19 for efficient secretion of glutathione. Also, Sinorhizobium fredii NGR234 lacks y-GCS as well as YbdK enzymes which makes it a good candidate for an introduction of glutamyl-cysteine ligase activity in its system.
Microorganisms, plasmids and primers used in this study
Bacteria and their growth conditions20 for this study are mentioned in Table 1. Plasmids and Primers used in this study are described in detail in Table 2.
Table (1):
Bacterial strains used in this study.
| Bacterial strain (Wild type /GMOs) and antibiotic selection | Growth medium | Optimum temperature | Source and description | |
|---|---|---|---|---|
| Wild type bacteria | Escherichia coli DH10B Streptomycin | Luria Bertani broth/agar (LB) | 37 °C | The department of Biochemistry, MSU Baroda |
| Sinorhizobium fredii NGR 234 Rifampicin | Yeast extract Mannitol broth /agar (YEM) | 28 °C | Generous gift by Dr. José Mª Vinardell González, Facultad de Biología, Universidad de Sevilla, Spain | |
| Transformants | Escherichia coli DH10B (pPAT) Kanamycin (25ug/ml) | Luria Bertani broth/agar (LB) | 37 °C | Escherichia coli DH10B transformed with the recombinant plasmid (pPAT) |
| Sinorhizobium fredii NGR 234 (pPAT) Rifampicin/Kanamycin(18ug/ml) | Yeast extract Mannitol broth /agar (YEM) | 28 °C | Sinorhizobium fredii NGR 234 transformed with the recombinant plasmid (pPAT) |
Genomic DNA extraction, plasmid purification and Transformation of bacteria
Genomic DNA from Escherichia coli DH10B was extracted by using a standard CTAB method.21 Plasmid purification was done by using a modified method involving silicone dioxide.22 Escherichia coli DH10B was transformed by a standard calcium chloride transformation method23 while Sinorhizobium fredii NGR234 was transformed by electroporation.24
Table (2):
Plasmids and primers used in this study.
Plasmid/Primers |
Size in base pairs (bp) |
Description |
Antibiotic selection |
Source |
|---|---|---|---|---|
pBBR1MCS2 |
5148 bp |
Low copy number cloning and expression vector having derepressed lac promoter. |
Kanamycin 25ug/ml |
The M.S. University of Baroda. |
pPAT-ybdK |
6264 bp |
1119 bp ybdK ligated in pBBR1MCS2 plasmid |
Kanamycin 25ug/ml |
ybdK gene was amplified by PCR using suitable primers from Escherichia coli DH10B genome and then ligated in pBBR1MCS2 plasmid after a restriction digestion of vector and PCR amplified gene |
Forward primer |
34 bp |
CGCTCGAGAGGAGGATAGTCATGCCATTACCCGA 34 base pairs primer flanking Xho1(CTCGAG) restriction enzyme site at 5’ end |
NA |
NA |
Reverse primer |
28 bp |
TTAAGCTTTTAGTCACCGGCCCAGATCT 28 base pairs primer flanking Hind3 (AAGCTT) restriction enzyme site at 5’end |
NA |
NA |
Glutathione estimation
Intracellular and extracellular glutathione was estimated by the method based on reduction of GSSG by NaBH4. This method depends on two measurements of GSH before and after reduction of GSSG in bacterial supernatant.25 Cell free lysate was used to estimate extracellular glutathione.
Detection of total intracellular ROS in bacteria by fluorescence microscopy and fluorometer
Sinorhizobium fredii NGR234 (pPAT) and Sinorhizobium fredii NGR234 cells were grown in YEM broth with the appropriate conditions mentioned above till their OD reaches 0.8. Approximately 1 ml cells were pelleted down at 8000 rpm for 2 minutes, washed with PBS twice and again resuspended in PBS for ROS determination. ROS was estimated by DCFDA which enters the cells and reacts with reactive oxygen to form green fluorescent colored compound di-chlorofluorescein (DCF).26 The estimation of ROS was done by both fluorometer and microscopy. A stock solution of DCF-DA (1mM) was prepared in PBS and was further diluted to 10 μM working concentration in the cells dispersed in PBS. Cells were then incubated for 30 minutes at 30°C in total dark condition. After that cells were again washed twice with PBS before fluorometry and microscopy experiment. Fluorescence intensity was measured at excitation of 485 nm and emission at 520 nm (Biotek). For both experiments unstained Sinorhizobium fredii NGR234 cells suspended in PBS were used as control. Microscopy was performed by fluorescent Nikon-Ti2E live imaging system at 60X magnification.
In vitro synthesis of Cadmium sulphide nanoparticles by wild type and genetically modified rhizobium
Bacterial cells were allowed to grow in YEM medium for 48 hr as mentioned in Table 1 and their cell free lysate was collected by centrifugation at 8000 RPM for 5 minutes, which was used for synthesis of cadmium sulphide nanoparticles. Cadmium chloride (0.25M) and sodium sulphide (0.25M) (1:1) were allowed to react with equal volume of cell free lysate at proper conditions to initiate the synthesis of cadmium sulphide nanoparticles.7
Characterization of Cadmium sulphide nanoparticles
First step in characterization of nanoparticles was to have a clearer look into its morphology by observing the biosynthetic nanoparticles under SEM (Sigma VP SEM/Carl Zeiss NTS) and recording their micrographs.7,27 FTIR spectrophotometry was performed by Bruker Alpha 2 spectrophotometer within the infrared range of 500-3500 cm−1, to have a better understanding of the functional groups attached with nanoparticles.28 Finally XRD was performed by XPERT PRO XRD machine to get a clear cut understanding about its crystalline nature. The intensities were recorded in a range of 10- 80° 2θ angle.7
Construction of a recombinant pPAT plasmid and transformation of bacteria
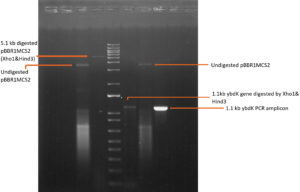
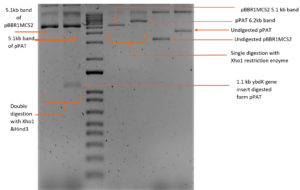
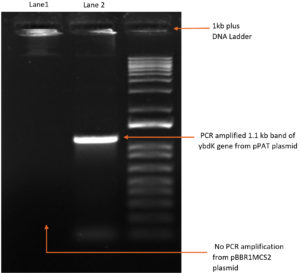
1119 bp ybdK gene was amplified from the genomic DNA of Escherichia coli DH10B by using forward and reverse primers flanked by Xho1 and Hind3 restriction sites respectively at their 5’ end (Table 3). Plasmid vector pBBR1MCS2 and ybdK gene (amplified from the genomic DNA) were sequentially digested with Xho1 and Hind3 restriction enzymes (thermofisher scientific)29 (Fig. 1a), followed by ligation using DNA ligase enzyme (thermofisher scientific). After ligation, Escherichia coli DH10B was transformed with ligation mixture and cells were spreaded on LB agar medium containing kanamycin 25µg/ml and allowed to grow overnight at 37°C. Single colony was selected from the transformation plate and allowed to grow in LB medium supplemented with kanamycin 25µg/ml till 0.8 OD. Cells were pelleted down and plasmid was isolated from them followed by its digestion with enzymes Xho1 and Hind3 giving rise to two distinct bands of 5.1 kb and 1.1 kb on 1% agarose gel, thus confirming the presence of recombinant plasmid pPAT in E.coli DH10B cells (Fig. 1b). Recombinant plasmid pPAT was purified from Escherichia coli DH10B and electroporated in Sinorhizobium fredii NGR234. 12 kv/cm at a fixed time of 5 ms gave maximum transformation efficiency. Recombinant plasmid was again isolated from Sinorhizobium fredii NGR234 and was confirmed by a PCR amplification of ybdK gene from the purified pPAT plasmid. PCR amplification gave a clear band of 1.1kb, which confirmed the transformation of Sinorhizobium fredii NGR234 by pPAT plasmid (Fig. 1c).
Table (3):
Extracellular and intracellular levels of glutathione produced by WT and GMO S.fredii NGR 234. All the values are represented in mean ± SD format.
Organism |
GSH mM |
GSSG mM |
Total Glutathione mM |
GSH/GSSG ratio of mean |
|
|---|---|---|---|---|---|
Extracellular |
Sinorhizobium fredii NGR 234 |
0.738 ±0.004 |
3.365± 0.035 |
4.07 ±0.001 |
0.21 |
Extracellular |
Sinorhizobium fredii NGR 234 (pPAT) |
0.375 ±0.021 |
93.835± 0.035 |
94.275± 0.035 |
0.004 |
Intracellular |
Sinorhizobium fredii NGR 234 |
0.650± 0.001 |
8.875± 0.021 |
9.520± 0.014 |
0.07 |
Intracellular |
Sinorhizobium fredii NGR 234 (pPAT) |
0.999 ±0.002 |
176.44± 0.015 |
177.445 ±0.008 |
0.005 |
[GSH- Reduced glutathione, GSSG- Oxidized glutathione]
Detection of total intracellular ROS in bacteria by fluorescence microscopy and fluorometer
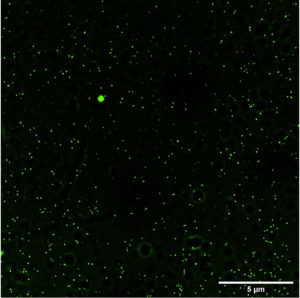
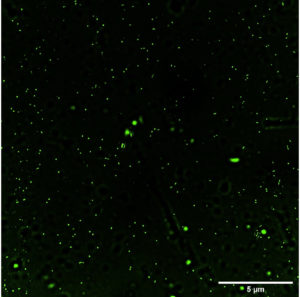
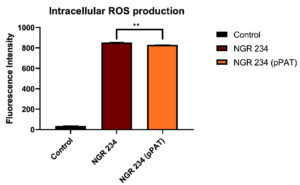
Fig. 2a and 2b (images taken by fluoroscent microscope) shows green coloured fluoroscence of DCF inside the bacterial cells. Equal number of Sinorhizobium fredii NGR234 and Sinorhizobium fredii NGR234 (pPAT) cells were taken and stained which showed that cloning Escherichia coli DH10B ybdk gene in Sinorhizobium fredii NGR234 significantly improved its oxidative status. Sinorhizobium fredii NGR234 (pPAT) showed less fluoroscence compared to Sinorhizobium fredii NGR234. For further confirmation fluoroscence intensity was measured by fluorometer which showed a significant decrease in fluoroscence emitted by GM bacteria compared to WT bacteria as shown in Fig. 2c. This is an indication of a proper heterologous expression of YbdK in Sinorhizobium fredii NGR234.
Fig. 2c. Intracellular ROS production by WT and GM bacteria. Data expressed as mean ± SEM, *p< 0.05, ** p<0.01 (n=3).
Bacterial growth curve
We examined whether an overexpression of YbdK enzyme has any effect on the growth of S. fredii NGR 234 or not. Sinorhizobium fredii NGR234 and Sinorhizobium fredii NGR234 (pPAT) were allowed to grow in YEM medium for 70 hours with proper conditions mentioned in Table 1 and OD at 600 nm was recorded every 4 hours. It was observed that S.fredii NGR 234 (pPAT) showed increased OD between 25th – 40th hour as compared to Sinorhizobium fredii NGR234 cells (Data not shown). This is due to increased production of glutathione which helps to reduce the total oxidative stress of the growth media.
Glutathione estimation
Intracellular and extracellular glutathione was estimated from Sinorhizobium fredii NGR234 and Sinorhizobium fredii NGR234 (pPAT). To estimate extracellular glutathione, cell free lysate was used. Cells and lysate were separately analyzed for glutathione after 48 hours incubation in YEM medium. 23 fold increase in extracellular levels of total glutathione (GSH+GSSG) was observed in Sinorhizobium fredii NGR234 (pPAT) compared to Sinorhizobium fredii NGR234 while intracellular total glutathione in Sinorhizobium fredii NGR234 (pPAT) increased upto 19 folds approximately compared to Sinorhizobium fredii NGR234 cells. Also Sinorhizobium fredii NGR234 (pPAT) showed 1.5 folds increase in the production of reduced GSH (Intracellular) as compared to Sinorhizobium fredii NGR234. Extracellular and intracellular GSH/GSSG ratio for Sinorhizobium fredii NGR234 was 0.21 and 0.07 respectively while the extracellular and intracellular GSH/GSSG ratio for Sinorhizobium fredii NGR234 (pPAT) was 0.004 and 0.005 respectively, mentioned in Table 3. Both Intracellular and extracellular total glutathione showed a significant increase in GMO bacteria compared to its wild type counterpart which clearly indicates the steady state constitutive expression of YbdK enzyme and in turn more production of glutathione. The drastic decrease in GSH/GSSG ratio of GMO bacteria compared to wild type bacteria could indicate more transport of GSH outside the bacterial cells, thus it has become clear that constitutive over expression of YbdK enzyme in Sinorhizobium fredii NGR234 can increase total glutathione levels inside and outside the cells. Moreover, it is a fact that glutathione reductase enzyme is only present inside the cell cytoplasm so the intracellular levels of GSH is higher in GMO bacteria compared to wild type. Higher levels of GSSG inside and outside the cells indicates an elevated production of GSH molecules which were later oxidized to GSSG and started accumulating. This is because the cells have basal levels of glutathione reductase enzymes while only YbdK enzyme was overexpressed, so this imbalance of alleviated glutamyl-cysteine ligase activity compared to glutathione reductase activity might be the reason for increased GSSG accumulation which is also an indirect sign for the proper overexpression of Escherichia coli DH10B YbdK enzyme in Sinorhizobium fredii NGR234. Due to lack of cognate antibodies for YbdK, western blot analysis was not performed.
Cadmium sulphide nano particle synthesis and characterization
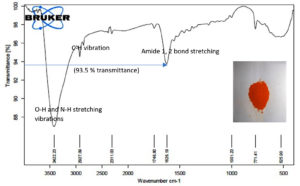
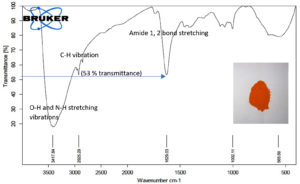
We tested the ability of Sinorhizobium fredii NGR234 (pPAT) cells to produce the nanoparticles. 50 ml cell free lysate of Sinorhizobium fredii NGR234 (pPAT) cells produced 2.059±0.001g cadmium sulphide bionanocrystals, while the cell free lysate of Sinorhizobium fredii NGR234 produced 2.009±0.001g cadmium sulphide bionanocrystals. We have seen that cell free lysate of S. fredii NGR 234 (pPAT) had a very high amount of GSSG (Table 3). Reaction between sodium sulphide and cell free lysate could have initiated a cascade of electron donation to GSSG which generated CdS nanoparticles.31 We also observed that the dry weight of the nanoparticles obtained from GMO (genetically modified) cells supernatant was about 20.28% more in comparison to the nanoparticles obtained from wild type cells supernatant. FTIR was performed further to get a clear idea about the functional groups attached with the nanoparticles. FTIR analysis showed various transmittance peaks between 3500cm-1 to 500cm-1 (Fig. 3a and b). The peaks at 1629.03 cm-1 and 3417.84 cm-1 for Sinorhizobium fredii NGR234 (pPAT) gave transmittance of about 51% and 19% respectively, while the similar ranged peaks at 1626.19 cm-1 and 3422.23 cm-1 from Sinorhizobium fredii NGR234 gave transmittance of 87% and 93.5% respectively. The decrease in transmittance of both peaks for Sinorhizobium fredii NGR234 (pPAT) suggests that it has successfully overexpressed glutathione. The peak around 1629 cm-1 in the Fig. 3a and 3b represents an amide 1 and 2 bonds stretching from the conversion of C-OH biomolecules. The amide bonds of protein and especially glutathione may interact with nanoparticles to stabilize it.30 Also the broad and strong peak between 3200 cm-1 – 3400 cm-1 indicates the stretching vibrations of O-H (phenols) and N-H (amines) bonds due to primary and secondary amine linkages of proteins and amino acids. Small peaks around 2927 cm-1 (Fig. 3a) and 2925 cm-1 (Fig. 3b) corresponds to the C-H vibration of alkanes. The data showed decrease in transmittance of the peak around 1629 cm-1 for Sinorhizobium fredii NGR234 (pPAT) compared to Sinorhizobium fredii NGR234. This corresponds to amide 1 and 2 stretching representing the increase in peptide bonds in the nanoparticles which indirectly confirms the elevated levels of glutathione in nanoparticles formed by Sinorhizobium fredii NGR234 (pPAT).
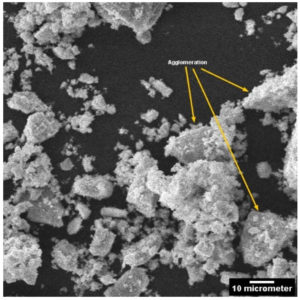
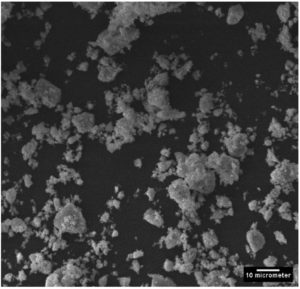
SEM observation revealed the capability of the organisms to produce variety of nanoparticles. The particles displayed vivid shapes ranging from a simple triangular shape to a complex heptahedron shapes for both organisms. Less aggregation and smaller size of nanoparticles were obtained by Sinorhizobium fredii NGR234 (pPAT) as compared to Sinorhizobium fredii NGR234 (Fig. 4a and 4b). It is already known that the organism which overexpresses glutathione tends to produce smaller and identical nanoparticles with very less aggregates and internal agglomeration.9 Average size of nanoparticles produced by Sinorhizobium fredii NGR234 was 909 nm while Sinorhizobium fredii NGR234 (pPAT) produced nanoparticles of size 343 nm suggesting the proper overexpression and secretion of glutathione outside the cells by GM bacteria. SEM images revealed that the nanoparticles formed by Sinorhizobium fredii NGR234 (pPAT) were comparatively smaller and showed less aggregation and showed better dispersion compared to the nanoparticles formed by Sinorhizobium fredii NGR234. Previously it has been reported that Escherichia coli ABLE C cells overexpressing glutathione produced more dispersed nanoparticles and showed less aggregation.9 It is possible that more number of glutathione molecules will bind more CDS particles thus sequestering smaller particles resulting in formation of dispersed nanoparticles. Such explanation has been previously stated by many researchers for the production of smaller nanoparticles by the microorganisms either overexpressing Phytochelatin or glutathione.11
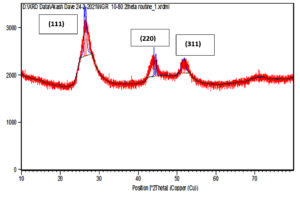
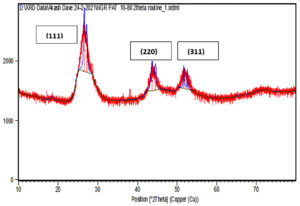
XRD data pattern of CdS nanoparticles from Sinorhizobium fredii NGR234 gave major peaks for 2 theta at 26.54, 44.56 and 51.86 angles which corresponds to (111), (220) and (311) planes (Fig. 5a). And the CdS nanoparticles from Sinorhizobium fredii NGR234 (pPAT) gave major peaks for 2 theta at 26.59, 43.67 and 51.65 angles which corresponds to (111), (220) and (311) planes (Fig. 5b). The lattice plane of CdS nanoparticles showed d spacing of 3.2 Å to the strongest peak (111) for the nanoparticles produced by both GMO and WT which indicates a face centered crystalline nanoparticle structure. The peaks obtained corresponds to the JCPDS card no. 10-454 for CdS nanoparticles. The main difference between the XRD peaks of nanoparticles produced by WT and GMO is the decrease in intensity of the peaks for S. fredii NGR 234 (pPAT) which indicates the smaller crystalline size compared to S. fredii NGR 2347. This results correlates with the SEM results where it was seen that S. fredii NGR 234 (pPAT) produces significantly smaller nanoparticles.
S. fredii NGR 234 is an agriculturally important rhizobium which exhibits a PGPR activity and can promote nitrogen fixation in multiple legumes. The aim behind this work is establishing S. fredii NGR 234 as a standard chassis for simple and complex genetic modification. Many rhizobium possesses multiple plasmids as large as 2-3 Mb in size which indicates that they can tackle plasmid load very easily. This ability should be put to use by introducing foreign genes which are industrially and agriculturally important to us. So we did this by introducing a plasmid containing ybdK gene in S.fredii NGR 234. It is evident from the above results that S. fredii NGR 234 is capable of synthesizing CdS nanoparticles. Also, it has become clear that overexpressing YbdK enzyme elevates glutathione secretion by S. fredii NGR 234 cells which in turn increase the synthesis of nanoparticles as well as change the physical and chemical properties of nanoparticles. Extracellular GSSG production by the genetic modified rhizobium was about 1.88 folds higher as compared to the wild type rhizobium. Freely available GSSG in cell free lysate would have acted as the precursor for the cascade reaction which starts as soon as sodium sulphide reacts with the cell free lysates. The sulphide would have acted as a nucleophile and donated its electrons to bisdisulphide of GSSG, similar to the transsulfuration reactions thus generating CdS and lanthi-MT type of molecules. So we conclude that more amount of GSSG in lysate can generate more thiolated molecules after a reaction with sodium sulphide thus decreasing the Cd/thiol ratio and generating smaller nanoparticles.
ACKNOWLEDGMENTS
The authors are very thankful to Dr. Jose Ma Vinardell Gonzalez, Facultad de Biologia, Universidad de Sevilla, Spain for donating S.fredii NGR234 strain to carry out this research work. Thanks to Prof. Rasheedunissa Begum & Prof. G. Naresh Kumar, Department of Biochemistry, M.S University, Vadodara for giving us an access to their lab facilities like Electroporator and Incubator shaker respectively. The authors also thank Department of Metallurgy and Engineering, M.S University, Vadodara for XRD analysis. GEER foundation, Gandhinagar for SEM analysis and Department of Chemistry, M.S University for FTIR analysis.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Briffa J, Sinagra E, Blundell R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon. 2020;6(9):e04691.
Crossref - Verma N, Sharma R. Bioremediation of Toxic Heavy Metals: A Patent Review. Recent Pat Biotechnol. 2017;11(3):171-187.
Crossref - Nanda M, Kumar V, Sharma DK. Multimetal tolerance mechanisms in bacteria: The resistance strategies acquired by bacteria that can be exploited to ‘clean-up’ heavy metal contaminants from water. Aquat Toxicol. 2019;212:1-10.
Crossref - Fagorzi C, Checcucci A, Dicenzo GC, et al. Harnessing rhizobia to improve heavy-metal phytoremediation by legumes. Genes. 2018;9(11):542.
Crossref - Iravani S. Bacteria in Nanoparticle Synthesis : Current Status and Future Prospects. Int Sch Res Notices. 2014;2014:359316.
Crossref - Hari S. Biosynthesis of nanoparticles from microorganisms. Res J Pharm Technol. 2020;13(4):2024-2028.
Crossref - Shivashankarappa A, Sanjay KR. Escherichia coli-based synthesis of cadmium sulfide nanoparticles, characterization, antimicrobial and cytotoxicity studies. Brazilian J Microbiol. 2020;51(3):939-948.
Crossref - Hamouda RA, Hussein MH, Abo-elmagd RA, Bawazir SS. Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Sci Rep. 2019;9(1):13071.
Crossref - Chen YL, Tuan HY, Tien CW, Lo WH, Liang HC, Hu YC. Augmented biosynthesis of cadmium sulfide nanoparticles by genetically engineered Escherichia coli. Biotechnol Prog. 2009;25(5):1260-1266.
Crossref - Yuan Q, Bomma M, Xiao Z. Enhanced silver nanoparticle synthesis by escherichia coli transformed with candida albicans metallothionein gene. Materials. 2019;12(24):4180.
Crossref - Yuan Q, Bomma M, Hill H, Xiao Z. Expression of Rhizobium tropici phytochelatin synthase in Escherichia coli resulted in increased bacterial selenium nanoparticle synthesis. J Nanoparticle Res. 2020;22:369.
Crossref - Choi Y, Lee SY. Biosynthesis of inorganic nanomaterials using microbial cells and bacteriophages. Nat Rev Chem. 2020;4(12):638-656.
Crossref - Yoon J, Shin M, Lim J, Kim DY, Lee T, Choi JW. Nanobiohybrid Material-Based Bioelectronic Devices. Biotechnol J. 2020;15(6):1-14.
Crossref - Choi Y, Park TJ, Lee DC, Lee SY. Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials. Proc Natl Acad Sci U S A. 2018;115(23):5944-5949.
Crossref - Kang SH, Bozhilov KN, Myung N V., Mulchandani A, Chen W. Microbial synthesis of CdS nanocrystals in genetically engineered E. coli. Angew Chemie – Int Ed. 2008;47(28):5186-5189.
Crossref - Monras JP, Diaz V, Bravo D, et al. Enhanced Glutathione Content Allows the In Vivo Synthesis of Fluorescent CdTe Nanoparticles by Escherichia coli. PLoS One. 2012;7(11):e48657.
Crossref - Rubino FM. Toxicity of glutathione-binding metals: A review of targets and mechanisms. Toxics. 2015;3(1):20-62.
Crossref - Lehmann C, Doseeva V, Pullalarevu S, Krajewski W, Howard A, Herzberg O. YbdK is a carboxylate-amine ligase with a γ-glutamyl: Cysteine ligase activity: Crystal structure and enzymatic assays. Proteins Struct Funct Genet. 2004;56(2):376-383.
Crossref - Schmeisser C, Liesegang H, Krysciak D, et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol. 2009;75(12):4035-4045.
Crossref - Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189(23):8746-8749.
Crossref - Atashpaz S, Khani S, Barzegari A, et al. A robust universal method for extraction of genomic DNA from bacterial species. Microbiol. 2010;79(4):538-542.
Crossref - Li JF, Li L, Sheen J. Protocol: A rapid and economical procedure for purification of plasmid or plant DNA with diverse applications in plant biology. Plant Methods. 2010;6(1):1-8.
Crossref - Chan WT, Verma CS, Lane DP, Gan SKE. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci Rep. 2013;33(6):e00086.
Crossref - Garg B, Dogra RC, Sharma PK. High-efficiency transformation of Rhizobium leguminosarum by electroporation. Appl Environ Microbiol. 1999;65(6):2802-2804.
Crossref - Alisik M, Neselioglu S, Erel O. A colorimetric method to measure oxidized, reduced and total glutathione levels in erythrocytes. J Lab Med. 2019;43(5):269-277.
Crossref - Pal K, Roy S, Parida PK, et al. Folic acid conjugated curcumin loaded biopolymeric gum acacia microsphere for triple negative breast cancer therapy in in vitro and in vivo model. Mater Sci Eng C. 2019;95:204-216.
Crossref - Sharma A, Kumar V, Kaur P, Thukral AK, Bhardwaj R. Phytochemical and elemental analysis of Brassica juncea L. leaves using GC-MS and SEM-EDX. Res J Pharm Technol. 2015;8(12):1662-1664.
Crossref - Hemashree T, Prasunna SG, Sakthiselvan P. Synthesis of Keratin Nanoparticle and Characterization using FTIR. Res J Pharm Technol. 2019;12(6):2664-2668.
Crossref - Ramos CRR, Abreu PAE, Nascimento ALTO, Ho PL. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal his-tagged fusion peptide. Brazilian J Med Biol Res. 2004;37(8):1103-1109.
Crossref - Wang SL, Chuang CH, Lin SY. pH-dependent coordination of metal-lisinopril complex investigated by attenuated total reflection/Fourier transform infrared spectroscopy. Chem Pharm Bull. 2002;50(1):78-82.
Crossref - Romero I, Tellez J, Yamanaka LE, Steindel M, Romanha AJ, Grisard EC. Transsulfuration is an active pathway for cysteine biosynthesis in Trypanosoma rangeli. Parasites and Vectors. 2014;7(1):197.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.