ISSN: 0973-7510
E-ISSN: 2581-690X
Bacterial blight in pomegranates (Xanthomonas citri pv. punicae (Xcp)), has become a severe concern in recent years. Hence, a molecular analysis of selected Xcp isolates and field samples from different locations was conducted. PCR amplification of the Xanthomonas Repetitive Intergenic Consensus (XRIC) box yielded 216 bp and 159 bp size bands in all Xcp as well as X. citri pv. citri (Xcc) isolates as well as infected field samples. The 16S rRNA gene and XRIC (216 bp) Xcp amplicons shared the highest sequence similarity with various Xcp and Xcc accessions. Two Xcp-specific SCAR primers were designed from in silico PCR simulation studies. SCAR Xcp1-20 and Xcp133-152 primers amplified a 152 bp band in all Xcp isolates (except one) as well as in infected pomegranate samples, while Xcc isolates yielded a 371 bp band. SCAR XcpF/R amplified a 200 bp band in all infected pomegranate samples as well as seven of eight Xcp isolates, while the Xcp Pune-Daund isolate, along with both Xcc isolates, amplified a 350 bp band. KM-gyrB primers amplified a 491 bp band in all Xcp isolates; and a 375 bp band in both Xcc isolates, as well as in 12 of 20 plant samples, with two yielding other-sized bands. Xac-rpf-specific primers amplified an expected 581bp band in all Xcp and Xcc isolates, as well as 14 out of 20 field samples. It can be concluded that the XRIC box amplification and sequencing method enables the rapid identification of various Xanthomonas species. Additionally, both SCAR primers developed can be directly used to identify the bacterial blight-infected field samples, facilitating rapid Xcp detection.
Molecular Diagnosis, Intergenic Hairpin-like Loops, Oily Spot
Pomegranate (Punica granatum L.) is a popular fruit crop grown both for juice and for table purposes. It has proved to be a blessing for dryland farmers of arid/semiarid regions due to its hardiness and versatile adaptability to thrive under low water availability. India contributes to 36 percent of the global production and 30 percent of the global pomegranate trade.1 Maharashtra State contributes to two-thirds of its pomegranate production. 1
Among the various diseases infecting pomegranate, bacterial blight caused by X. citri pv. punicae (Xcp) is a serious threat due to its high epidemic potential.2,3 It was of minor importance until the early 1990s, when it reached epidemic proportions, resulting in yield losses of 60-80 percent.4 A further outbreak of this disease has been severe and has acquired an epidemic status in the pomegranate belt of peninsular India.5,6 Later on, it was reported outside of the Indian subcontinent.7,8
The disease infects various aerial plant parts including pomegranate fruits which have lower market value and are not suited for export purposes due to strict quarantine regulations. On average, this disease causes 30%-50% losses, with up to 80% losses reported under epiphytotic conditions. Orchard owners are required to destroy the disease-infected plants and orchards. Species of Xanthomonas are difficult to eradicate once they infest any area and strict quarantine measures need to be implemented. Therefore, clean cultivation, effective diagnosis and management are recommended.
The traditional identification of Xanthomonas includes isolation, purification, and morphological-cultural studies,9 but some strains of this genus are reportedly difficult to isolate. Classical methods have a few shortcomings, including a lack of sensitivity and specificity. Molecular diagnostic approaches have been used both at the laboratory and field levels. Pathogen-specific primers have been used for the rapid identification of many phytopathogenic bacteria, including Xanthomonas with initial reports from the 1990s.10-12 Plasmid restriction digestion, 16S rRNA gene sequences, intergenic spacer regions, enterobacterial repetitive intergenic consensus (ERIC), repetitive element sequence-based PCR (REP-PCR), box-based PCR, and gene-specific primers have already proved their importance in diversity studies and diagnostic applications in bacterial diagnosis.13-16 On the molecular evaluation of Xcp isolates, their variability was found to be independent of geographic allocation.17
XRIC (Xanthomonas Repeat Intergenic Region) are imperfect palindromic repeats, initially identified from genes involved in xanthomonadin pigment biosynthesis.18 By evaluating sequence information available on the web, bioinformatics tools based on in silico PCR simulations can be used efficiently to design primers specific for molecular diagnosis and pathogen detection. However, PCR primers thus designed for diagnosis need to be validated both using pure bacterial cultures and infected plant samples. This study was undertaken for the validation of Sequence Characterized Amplified Regions (SCAR) primers designed from the XRIC sequence, along with a comparison with previously reported related primers in a larger sample size. This method allows for the early detection of phytopathogens such as Xanthomonas, in nurseries. The aim of this study was to develop and validate PCR primers specific to Xcp as pure isolates as well as bacterial blight-infected samples.
Collection of pathogen samples
A survey was conducted in the pomegranate-growing regions of western Maharashtra, including Solapur, Nashik, Pune, and Ahilyanagar districts, to collect bacterial blight-infected pomegranate samples (Table 1). Plants were confirmed as infected based on typical symptoms. Xanthomonas were isolated from macerated, infected pomegranate samples, streaked on nutrient agar medium with sucrose (NAS) and further incubated at 28 ± 2 °C for 48-72 h.19 After 48-72 h they were examined for the presence of typical colonies, which were transferred to NAS slants and maintained on yeast extract glucose agar with a charcoal slant for further studies. Eleven pure isolates of X. citri pv. punicae (Xcp) grown on nutrient agar slants were further used for molecular characterization. These bacterial isolates were maintained on YEM agar slants at 4 °C.
The details of these isolates collected from Western Maharashtra are given in Table 1. A pure isolate from Deola-Nashik was used as a positive control for the confirmation of field samples. Simultaneously, crude extracts of naturally bacterial blight-infected pomegranate samples, as well as artificially inoculated plants, were also used along with two X. citri pv. citri (Xcc) and six isolates of X. vesicatoria from different parts of Maharashtra (from another study) for comparative molecular studies. Eleven Xcp bacterial isolates were initially evaluated using biochemical tests, viz. Gram staining, KOH solubility, Kovac’s oxidase, starch hydrolysis, lipase activity, arginine dihydrolase test, gelatin hydrolysis, and catalase tests, as well as for growth on 0.2% asparagines medium. A pathogenicity test was conducted using the standard syringe infiltration method on leaves of one-year-old pomegranate cv. “Ganesh”. Freshly grown (48 hrs, 28 °C) bacterial suspension (107 to 108 CFU/ml) was syringe infiltrated into recently expanded leaves.
Table (1):
Bacterial samples used for PCR studies
No. |
Species/pathovar |
Details of geo-graphical origin |
|---|---|---|
1 |
X. citri pv. punicae (Xcp) |
Ahilyanagar |
2 |
X. citri pv.punicae (Xcp) |
Solapur |
3 |
X. citri pv. punicae (Xcp) |
Solapur-Akkalkot |
4 |
X. citri pv. punicae (Xcp) |
Solapur-Pandharpur |
5 |
X. citri pv.punicae (Xcp) |
Ahilyanagar-Sangamner |
6 |
X. cirti pv.punicae (Xcp) |
Nashik-Deola |
7 |
Unpurified culture |
Ahilyanagar |
8 |
Unpurified culture |
Ahilyanagar |
9 |
Unpurified culture |
Ahilyanagar |
10 |
X. citri pv punicae (Xcp) |
Ahilyanagar |
11 |
X. citri pv. punicae (Xcp) |
Pune (Daund) |
12 |
X. citri pv.citri (Xcc) |
Nagpur |
13 |
X. citri pv.citri (Xcc) |
Tamilnadu |
Collection of infected pomegranate plant samples for diagnosis
Bacterial blight/oily spot disease-infected pomegranate plant samples were collected separately from the orchards of different pomegranate growing areas of district Nashik (Maharashtra state) i.e., Deola, Malegaon, Satana and Kalwan Tehsils (Table 2). The plant samples taken were freshly infected pomegranate plant leaves showing water-soaked oily coalesced lesions. The collection was done in the early symptoms stage. The infected plant parts were collected from orchards, wrapped in aluminum foil, labeled, and kept in liquid nitrogen till genomic DNA isolation in the laboratory for further studies.
Table (2):
Infected plant samples used for PCR studies
Sample No. |
Details of bacterial blight infected plant samples studied |
|---|---|
#1 to #6 |
Deola, Nashik District; leaves with water-soaked lesions |
#7 to #11 |
Satana, Nashik District; leaves with water-soaked lesions |
#12 to #15 |
Malegaon, Nashik District; leaves with water-soaked lesions |
#16 to #20 |
Lakhmapur village (Satana, Nashik District), Leaves with water-soaked lesions |
#21 |
Deola, Nashik District |
#22 |
Kalwan, Nashik District |
#23 |
MPKV, Rahuri, Ahilyanagar District |
Genomic DNA isolation from Xanthomonas pure bacterial cultures
In this investigation, bacterial genomic DNA was isolated from three-loops full of four to five days old culture grown on nutrient agar slants using the Sarkosyl 2% Protease digestion method.16 It involved dissolving culture pellets in sarkosyl 2% (dissolved in suspension buffer 50 mM Tris, 20 mM EDTA) and 7.5 µl of predigested protease and incubating at 50 °C until the solution had cleared. Phenol-chloroform extraction was followed by isopropanol precipitation of DNA and suspended pellets in 50 µl of TE buffer (10 mM Tris and 1 mM EDTA). In the case of crude extracts, infected tissue debris was removed by brief a spin (10,000 rpm for 3-5 minutes) to avoid blockage of the spin column.
Plant Genomic DNA isolation from infected leaves of pomegranate
In this investigation, twenty bacterial blight-infected pomegranate leaf samples (100 mg each) were homogenized with liquid nitrogen in sterile mortars and pestles, and plant genomic DNA was isolated using HiPurA™ Plant DNA Miniprep Purification kit.
The genomic DNA was quantified on Nanodrop Spectrophotometer (ND-1000) at 260 nm. The genomic DNA band intensity in a 0.8% agarose gel was visualized along with that of standard Lambda phage DNA (50 ng) on a Fluor Chem.TM Alpha Innotech gel documentation unit.
Cloning and sequence analysis of eluted fragments
XRIC (216 bp) and 16S rRNA (1500 bp) DNA fragments from a strain from Deola (District-Nashik) were eluted from the gel, cloned into a PCR cloning vector pTZ57R/T (2886 bp), and transformed into competent Escherichia coli strain JM109. Custom Sanger sequencing was done for cloned fragments in a vector. Manual sequence editing was done using the sequence analysis tools (ChromasLite 2.01 software) and the sequence results were analyzed by BLAST analysis. After sequence annotations, the details were submitted at www.ncbi.nlm.nih.gov as BankIT submissions with GenBank accession numbers viz., FJ827773.1 (for XRIC hairpin loop-forming region genomic sequence) and FJ827774.1 (for 16S-23S ribosomal RNA gene region with ITS spacers).
Computational analysis
Sequenced data from the XRIC region (acc. no. FJ82777) were assessed for all computational analyses. Web-tool Primer3 was used to design the two SCAR primer pairs (XRIC-Xcp_F/R) (F-Primer: 9-28 bp 5′-CAAAACTTACTGCGCAACCA-3′ and R-Primer: 187-206 bp 5′-CTAACAAAACGGAGCGAGCA-3′) and Xcp1-20F + Xcp133-152R (Xcp1-20F-primer GCGGCTAACAAAACTTACTG and Xcp133-152R Primer CCACTGTACGCATCAGATAG).20 These SCAR primer sets and the XRIC-Box primer, were initially used for in silico PCR amplification studies. This in silico amplification simulation experiment was carried using the online available prokaryotic genomes sequence data as templates at the site http://insilico.ehu.es/PCR/.21 For checking amplification possibilities, the genomes of five individual genera from the order Xanthomonadales, as well as twenty other common genera of class Gammaproteobacteria and 40 genome accessions of the genus Bacillus (as negative control) were analyzed. The online-generated virtual banding patterns were observed for their uniqueness and applicability in diagnostic applications.
PCR analysis of both bacterial/plant genomic DNA samples
Genomic DNA amplification of both bacterial and plant genomic DNA samples was carried out by PCR amplification in an Eppendorf Master Cycler. The custom synthesized primers used for the PCR amplification in the current investigation are listed in Table 3. Using gradient PCR amplification, the annealing temperature of primer pairs i.e., Xcp1-20+Xcp133-152, Xac01. rpf+Xac02.rpf, Xoo.TXT+Xoo.TXT.4R, Xcp-F+Xcp-R, and KM.gyr-5F+KM.gyr-6R was optimized.
Table (3):
PCR primer used for the amplication studies
| No. | Primer | Sequence 5’ to 3’ | Reference |
|---|---|---|---|
| 1 | 16F27 | AGAGTTTGATCMTGGCTCAG | Hauben et al.23 |
| 16R1525 | TTCTGCAGTCTAGAAGGAGGTGWTCCAGCC | 16S rRNA specific | |
| 2 | XRIC | AGAGCGGCTAACAAAACG | Goel et al.18 |
| 3 | Xcp-F | CAAAACTTACTGCGCAACCA | SCAR Current investigation |
| Xcp-R | CTAACAAAACGGAGCGAGCA | ||
| 4 | Xcp1-20 | GCGGCTAACAAAACTTACTG | SCAR Current investigation |
| Xcp133-152 | CCACTGTACGCATCAGATAG | ||
| 5 | KM.gyr-5 F | GTTGATGCTGTTCACCAGCG | Mondal et al.24 |
| KM.gyr-6R | CATTCATTTCGCCCAAGCCC | ||
| 6 | Xac01.rpf | CGCCATCCCCACCACCACCACGAC | Coletta-Filho et al.25 |
| Xac02.rpf | AACCGCTCAATGCCATCCACTTCA | ||
| 7 | Xoo.TXT.F | GTCAAGCCAACTGTGTA | Sakthivel et al.26 |
| Xoo.TXT.4R | CGTTCGCGCCACAGTTG |
The purified genomic DNA extracts of all the genotypes were used as template DNA. The amplification reaction mixture of 20 µl volume comprised of 1-unit Taq DNA Polymerase, 1X Buffer B (without MgCl2), 1.2 mM MgCl2, 1.0 mM dNTP, 10 picomoles of each primer, 30 ng template DNA and water. No template DNA was added in a total negative control PCR reaction.
For PCR amplification, the thermal profile comprised of initial denaturation for five minutes at 94 °C, followed by a PCR regime comprising of 40 cycles of denaturation (94 °C), annealing (50-61 °C gradient) and extension (72 °C) for one minute each, followed by final extension for 10 minutes at 72 °C and finally held at 4 °C till samples were retrieval. After standardization of annealing temperature, the amplification was carried out again to get prominent and specific bands, i.e. 16F27 + 16R1525, 16S RNA-specific (61 °C), XRIC (61 °C), Xcp1-20 + Xcp133-152 (55 °C), XcpF + XcpR (55 °C), KM.gyr5F + KM.gyr-6R (55 °C), Xac01rpf. + Xac02.rpf (55 °C) and Xoo.TXT + Xoo.TXT (56 °C), were used with others for the PCR described before.
The PCR products were resolved by 2% agarose gel electrophoresis along with a GeneRulerTM 100 bp DNA ladder. Gel electrophoresis was conducted at 80 volts for one to two hours and bands were visualized and documented in a gel documentation system. PCR profiles were visualized in the gel documentation unit.
Molecular diagnosis approaches have widescale applications in detecting pathogens alongside conventional diagnosis methods.22 They can be used for the successful detection of Xanthomonas from symptomatic as well as latent infection in host tissues. Detection of symptomless propagation material is of importance because latent infection populations can lead to disease development and severe epidemics under favourable conditions. In the present investigation, a survey was undertaken to detect the pathogen causing bacterial blight (oily spot) in pomegranates by using bacterial pathogen-specific primers.
Microbial and biochemical tests
Microbial phenotype and biochemical characterization of the collected bacterial isolates revealed that the pathogenic bacteria were Xanthomonas spp. The collected isolates when grown on NA at 28 °C had smooth, round, yellow-coloured colonies with entire margins (Figure 1a). They were Gram-negative bacteria as they gave pinkish staining with counter stain safranin; which was confirmed by string formation of the culture when using 3% KOH test. All eleven Xcp isolates were weakly positive in the oxidase test, positive in the catalase test, hydrolyzed starch as indicated by a clear zone around colonies when iodine stained media was used and showed gelatin liquefaction. Similar test results were earlier reported in Xcp16 and Xcc27, they tested negative for the arginine dihydrolase test: as they turned to the orange-pink colour in the presence of the pH indicator phenol red. No growth was observed on asparagine medium; typical of the presence of Xanthomonas; which is a characteristic feature that distinguishes it from closely related pseudomonads.28
Pathogenicity test
The Xcp isolates were shown to be pathogenic. After artificially inoculating the pomegranate plants, typical water-soaked angular lesions appeared after 7 days. Isolation of the pathogen from artificially infected plants proved Koch’s postulate, thereby confirming their pathogenicity. Sharma et al. evaluated various Xcp inoculation methods on pomegranates, of which the spray method yielded the most reproducible symptoms.29 The host specificity of Xanthomonas species or pathovars is generally limited.30
Molecular characterization
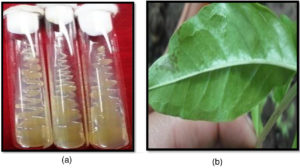
Genomic DNA isolated from thirteen isolates of Xanthomonas species [including eleven of Xcp from Maharashtra and two Xcc isolates (from Tamil Nadu and Nagpur)] as well as twenty infected leaf samples [from four locations of Nashik district, Maharashtra, India] were diluted to a uniform concentration of 10 ng/µl for use as PCR templates (Figure 1a and b).
Figure 1. a) Pure culture of Xcp on nutrient agar medium used for genomic DNA isolation; b) Bacterial blight infected leaf samples used for DNA isolation
In the present study, molecular detection of the bacterial pathogen was undertaken using different primers, specifically SCAR-based markers from the Xanthomonas repetitive intergenic consensus (XRIC) sequence as well as those reported previously in literature for Xcp and related Xanthomonas. Six of the seven primers amplified the products.
16S ribosomal RNA gene-specific PCR amplification studies
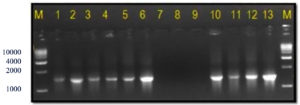
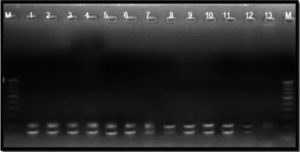
The 16S rRNA gene-specific primer pair (16F27 + 16R1525) amplified a 1500 bp fragment in all bacterial samples (except the three unpurified cultures of bacterial samples numbers 7 to 9) (Figure 2) and among 17 of 20 bacterial blight-infected plant DNA samples (except plant samples numbers 2, 4, and 19) (Figure 3) confirming the presence of bacteria.
Figure 2. Bacterial genomic DNA amplified using 16S rRNA primers
M: 1 Kb DNA ladder (GeneRulerTM); Lanes 1 to 6, 10 and 11: Xcp DNA amplified product; Lanes 7 to 9: Xcp Unpurified culture DNA amplified products; Lanes 12, 13: Xcc DNA amplified products
Figure 3. Infected plant DNA amplified using 16S rRNA primers
M: 1 Kb DNA ladder (GeneRulerTM); Lane 1 to 20: Infected plant DNA amplified products
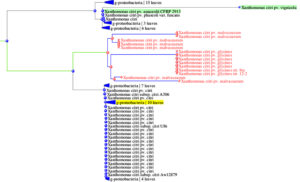
Initially, a 1500 bp 16S rRNA gene band from single isolate Xcp Deola-Nashik was cloned and sequenced and a 1018 bp sequence was obtained (GenBank accession FJ827773.1). In a sequence similarity analysis using NCBI megablast (Highly similar sequences), 100 BLAST hits were observed with 98%-99% coverage and 98.53%-99.51% identity. These BLAST results were further subjected to Distance tree results by selecting the Maximum Fast Evolution Method. It revealed that the closest accessions were six Xcp strains; which together with 32 Xcc accessions formed a subcluster (Figure 4). Another subcluster was comprised of seven X. citri pv. malvacearum (Xcm) and seven X. citri pv. glycines (Xcg) accessions. This high sequence identity observed in the 16S rRNA gene suggests that Xcp and Xcc are closely related to Xcm and Xcg. Another close X. citri cluster comprised of accessions of X. pv. phaseoli; X. pv. vignifola, and X. pv. perforans.
16S rRNA gene (1500 bp) sequencing has been extensively used for bacterial characterization and it can assist with bacterial taxonomic resolution.31-33 Earlier the same primers were used to establish the phylogenetic relationships in the genus Xanthomonas which had high sequence similarity (98.2%) of the 16S ribosomal RNA gene.23 High sequence similarity (95%-98%) was also reported in the 16S-23S rDNA intergenic region between X. oryzae pv. oryzae and X. campestris pathovars.34
Extensive molecular studies have reviewed the classification of Xanthomonas based on the genomic diversity and relationships therein.35,36 X. citri have been grouped into the subspecies Xcc and Xcm (both included pathovars from diverse hosts).37 It was proposed that Xcp is very close to X. citri subsp. malvacearum.38 Recently, genome sequences (4.94 Mb) of Xcp were found to have 98.78% to >99% nucleotide identity with X. citri pv. citri.39,40
XRIC (Xanthomonas Repeat Intergenic Consensus) box amplification and sequence study
Previously, a 145 bp XRIC sequence Box was reported, and was suggested to be Xanthomonas specific, and that over a hundred copies of the box may be present in the genome in immediate upstream or downstream regions of various genes.18 The XRIC box sequence formed a hairpin-like loop structure, with multiple copies including those in the pigmentation locus. They further suggested that the 64% G+C content of these XRIC elements was characteristic of Xanthomonas. Previously repetitive extragenic palindromic sequence PCR was used for the diagnosis of X. euvesicatoria.15
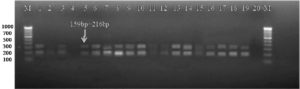
During the current investigation, a uniformly monomorphic pattern: with twin fragments of sizes 159 bp and 216 bp were observed in eight Xcp pure isolates and two Xcc isolates, as well as in twenty infected field samples. The 159 bp and 216 bp product lengths were observed in all three unpurified bacterial cultures (Figure 5). Among the twenty infected plant samples, eighteen of them amplified 159 bp and 216 bp bands, but samples no. 2 and 20 did not amplify (Figure 6). Similar profile was also observed in two crude extracts of naturally infected and the artificially inoculated plant sample (Figure 6). Amplification was not observed in either of the negative controls, i.e., healthy plant DNA or without template DNA. The amplification profile of X. vesicatoria isolates varied both in the number of amplicons as well as their polymorphic profile, with their size ranging from 180 bp to 700 bp.
Figure 5. Bacterial genomic DNA amplified using XRIC primer
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 6, 10-11: Xcp DNA amplified products; Lanes 7 to 9: Unpurified culture DNA; Lanes 12-13: Xcc DNA amplified products
Figure 6. Bacterial blight infected plant DNA amplified using XRIC primer
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 20: Infected plant DNA amplified products
PCR amplification derived 216 bp XRIC band was cloned and sequenced (GenBank accession #FJ827773). In the initial sequencing study, only a single Xcc (Strain 603) accession (Hairpin loop forming Xanthomonas specific region) showed a significant match. However, during subsequent sequence identity studies using NCBI megablast (Highly similar sequences), fifty-four BLAST hits were observed with 98%-100% coverage and 94.91%-97.18% identity. These 54 similar accessions included 33 sequence accessions from Xcc, seven from Xcp strains, seven from Xcg, six from Xcm, and one unclassified Xanthomonas. This indicates that the Xcp, Xcc, and Xcm accessions are closely related. In the same analysis (highly similar BLAST), another 32 accessions showed limited sequence similarity with the 3’end region of this accession (for the last 162-216 bp region), with 12%-25% coverage and 85.7%-100% identity; with 18 of those accessions being X. oryzae strains and the other 14 accessions being X. campestris. The option of somewhat similar sequences (BlastN) yielded a total of 100 BLAST hits with a complete 100% coverage and 76.44%-96.76% identity in various Xanthomonas.
Thus, in the present study, the XRIC sequence (accession no. FJ82777) from an Xcp isolate revealed that the initial 170 bases were conserved/shared only with closely related Xcp, Xcc, Xcg and Xcm; while in contrast, the last 50 bases of the accession were shared by various other Xanthomonas. This suggests that the initial 170 bp region is a good candidate for designing Xcp specific primers. As a result, it was proposed that the XRIC based PCR diagnosis can be used in the identification or detection of Xanthomonas in field diseased plant samples. Most of the conserved sites were intergenic regions either related to genes encoding pathogenesis-related or hypothetical proteins.
During an in silico simulation study of the available database using available prokaryotic genomes, none of the 25-genus other than Xanthomonas yielded any amplification, even under less stringent conditions with XRIC primers (results are not presented). These twenty-five-genera included five genera of order Xanthomonadales, i.e., Stenotrophomonas; Xylella; Pseudoxanthomonas; Frateuria, and Dyella, as well as common pathogenic bacteria like Pseudomonas, Erwinia, and Ralstonia. Under stringent conditions simulation (no mismatch allowed), XRIC box primer in silico amplification was observed in seven species/pathotypes amongst the panel of 14 Xanthomas; including three Xcc strains (amplified a 144 bp amplicon); three X. oryzae pv. oryzae (128 bp and 144 bp amplicons) and a X. oryzae pv. oryzicola (125 bp and 1156 bp amplicons). However, under less stringent in silico amplification conditions (two base mismatches allowed with none at the 3’ end), all 14 species/pathotypes of Xanthomonas yield amplification, with all of them yielding multiple bands, including common amplicons in the range of 142-146 bp.
SCAR based Xcp primers for specifically detecting Xcp
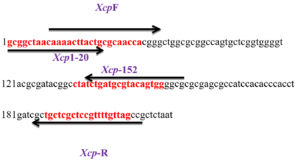
Based on these findings, that this region is highly pathovar specific, two new pairs of SCAR primers, Xcp1-20 + Xcp133-152 and XcpF + XcpR, were designed using Primer3 software at the website www.ncbi.nlm.nih.gov (Figure 7). Xcp1-20 + Xcp133-152 was designed to amplify the 152 bp region by excluding the ~60 bp terminal region shared with different Xanthomonas species; while XcpF/R was designed to amplify the almost complete region. Further, in silico PCR amplification under stringent conditions using both newly designed SCAR primer sets revealed no amplification in any other member of Xanthomonas (including three Xcc strains among the panel of 14 members) or Pseudomonas. With XcpF + XcpR primers, under less stringent conditions (two mismatches allowed with one in the last 3 nucleotides at the 3’ end): an amplicon of size 129 bp was observed in only two members of Xoo (KACC10331 and MAFF 311018). The size of those two amplicons observed in this study was significantly smaller than that of 129 bp specific to Xcp.
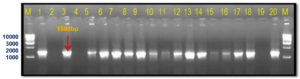
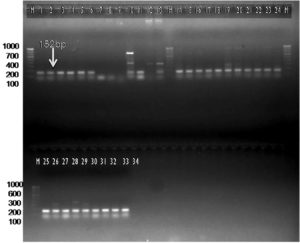
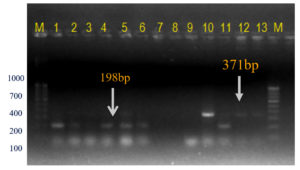
The current investigation was undertaken to further confirm/validate the in silico amplification results by further in vitro PCR amplification with the designed primer sets. The newly designed SCAR-based primer pair Xcp1-20+Xcp133-152 amplified a 152 bp band in all Xcp isolates and bacterial blight-infected pomegranate samples (Table 4; Figure 8). In no. 10 Pune-Daund Xcp isolate, additional 371 bp and 720 bp bands were amplified. In the two Xcc isolates (Tamil Nadu and Nagpur isolates) (no. 12 and no. 13) yielded 371 bp bands. Three unpurified cultures, 7, 8 and 9, lacked amplification, likely due to the absence of Xanthomonas. All the twenty-disease infected pomegranate leaf samples used for the amplification amplified a 152 bp product (Figure 8).
Table (4):
Detailed PCR amplification pattern
| No. | Primer | Infected plant sample | Xcp strain | Xcc strain | Exception | Unpurified strain |
|---|---|---|---|---|---|---|
| 1 | XRIC | 159 bp and 216 bp | 159 bp and 216 bp | 159 bp and 216 bp | – | 159 bp + 216 bp |
| 2 | SCAR XcpF+R | ~200 bp | ~200 bp | 350 bp | 350 bp (Pune-Daund) |
No amplification |
| 3 | SCAR Xcp120 + 133-152 | 152 bp | 152 bp | 371 bp | 152 bp, 371 bp, 720 bp (Pune-Daund) | |
| 4 | Xac01 + Xac02 | 581 bp | 581 bp | 581 bp | – | |
| 5 | KM-gyrase | 491 bp | 491 bp | 375 bp | 375 bp, 1088 bp (#8 and 18) | |
| 6 | 16SF27 + 16SFR1525 | 1500 bp | 1500 bp | 1500 bp | – | |
| 7 | Xoo.TXT + Xoo.TXT4R | No amplification | No amplification | No amplification | – |
Figure 8. Bacterial and plant DNA amplified using SCAR Xcp1-20 + Xcp133-152 primer pair
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 6, 10-11: Xcp DNA amplified products; Lanes 7 to 9: Unpurified culture DNA; Lanes 12-13: Xcc DNA amplified products; Lanes 15 to 34: BB infected plant DNA amplified products
Another SCAR Xcp F/R primer pair amplified a 200 bp band in all blight-infected plant samples, as well as seven of eight Xcp, isolates collected from western Maharashtra. Pune-Daund Xcp isolate no. 10 amplified a 350 bp band along with both Xcc isolates nos. 12-13, while three unpurified bacterial cultures did not amplify (Table 4; Figure 9). All twenty infected pomegranate DNA samples amplified a 200 bp band with primer pair XcpF + XcpR (Figure 10). Under in silico conditions, the pathovar-specific primer set, Xcp-F/R and Xcp1-20 + Xcp133-152 didn’t amplify in any other Xanthomonas except Xcc.
Figure 9. Bacterial genomic DNA amplified using SCAR XcpF + XcpR primer pair
M: Low range 100 bp plus DNA ladder (GeneRulerTM); Lanes 1 to 6, 10-11: Xcp DNA amplified products; Lanes 7 to 9: Unpurified culture DNA; Lanes 12-13: Xcc DNA amplified products
Figure 10. Infected plant DNA amplified using SCAR XcpF + XcpR primer pair
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 20: Infected plant DNA amplified products; Lanes 1 to 20: Infected plant DNA amplified products
Previously seven Xanthomonas effector xop genes were evaluated for diagnosis, of which xopQ primers yielded a 190 bp amplicon in plants infected with Xcp and Xcp.41,42 A loop-mediated isothermal amplification (LAMP) technique of Xcp was developed and validated in pomegranates using various sets of 16S rRNA primers.43
Verification of previously reported gyrase (gyrB) gene-based primer for detecting Xcp
Previously, the gyrB gene was used for identifying Xanthomonas species.24 They reported that Xcp isolates amplified a 491 bp band. Similarly, a specific 491 bp gyrB amplicon was also reported in bacterial blighted (Xcp) infected field samples of pomegranate.6 The identity of the Xap isolates was confirmed by specific gyrB and 16S rRNA gene sequences, with all isolates resulting in amplicon sizes of the PCR products were 491 bp and 1450 bp respectively.33 Previously phylogenetic analysis of 203 isolates of Xanthomonas was done using their gyrB gene sequences.36
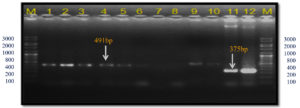
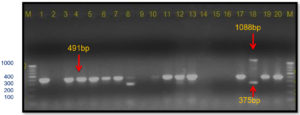
In the present study, these gyrB gene-specific primers KM gyr-F/R amplified a 491 bp band size in all Xcp isolates; however, in both Xcc isolates (no. 11 and 12), a 375 bp band was observed (Table 4; Figure 11). In the unpurified cultures no. 7-9, there was no amplification. The 491 bp was also observed in 12 out of 20 field samples. However, in two samples from the Satana, different bands were observed, i.e., sample #8 (320 bp) and #18 (491 bp and 1088 bp bands) (Figure 12).
Figure 11. Bacterial genomic DNA amplified using KM.gyr primer pair
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 6, 9-10: Xcp DNA amplified products; Lanes 7, 8: Unpurified culture DNA; Lanes 11-12: Xcc DNA amplified products
Figure 12. Infected plant DNA amplified using KM.gyr primer pair M: 100 bp DNA ladder; Lanes 1 to 20: Infected plant DNA amplified products
Verification of previously reported Xac.rpf primers
Previously, primers were developed for the detection of Xcc derived from the rpf gene region, in citrus plants.25 They reported that a 581 bp band was amplified in all Xcc infected samples. From the previous sequence similarity studies conducted in our laboratory it was observed that X. citri pv. citri was the closest pathovar, specifically in terms of sequence identity to X. citri pv. punicae. The primer set for the ITS of ribosomal RNA gene sequences was highly specific for X. citri pv. citri, whereas the pthA gene primer set was common to all strains of citrus blight canker Xanthomonas.13 On the contrary, leucine-responsive regulatory protein (lrp) gene was informative in distinguishing groups of Xcc.44 It was proposed that citrus canker causing Xanthomonas has self-mobilizing plasmids with pathogenicity factors, capable of transferring pathogenicity into other xanthomonad residents on the same citrus plant.45 A highly reproducible ligation-mediated PCR technique was developed based on three insertion sequences (IS-LM-PCR) in Xcc.46
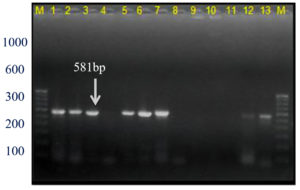
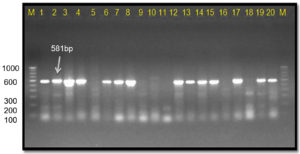
This Xcc-specific Xac.rpf primer pair amplified a 581 bp product in almost all the Xcp/ Xcc bacterial isolates across various geographical locations, matching the previously reported size (except for three unpurified culture nos. 10, 11, and 12 from the Ahilyanagar region) (Table 4; Figure 13). It also amplified in 14 of 20 bacterial blight-infected pomegranate plant samples, yielding a 581 bp product; while six samples were from Deola and Satana tehsils (no. 6, 10, 11, 12, 17 and 19) did not amplify (Figure 14). These results in the current investigation matched expected sequence similarity studies. However, the Xac.rpf primers are not specific to Xcc only but they are also amplified in Xcp and could not differentiate them from Xcc either.25
Figure 13. Bacterial genomic DNA amplified using Xac01+Xac02 primer pair
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 6, 9-10: Xcp DNA amplified products; Lanes 7, 8: Unpurified culture DNA; Lanes 11-12: Xcc DNA amplified products
Figure 14. Infected plant DNA amplified using Xac01 + Xac02 primers
M: 100 bp DNA ladder (GeneRulerTM); Lanes 1 to 20: Infected plant DNA amplified products
Verification using primer for specifically detecting X. oryzae pv. oryzae
Xanthomonas oryzae pv. oryzae (Xoo) based primers developed by earlier workers26 were included in the current studies as the in silico PCR simulation studies with Xcp primers had shown amplicons under less stringent conditions (2 mismatches) though of varying sizes in Xoo. Insertion sequence IS1112 based PJEL1/2 primers were also used to generate specific and reproducible PCR fingerprint patterns for the Xoo isolates.47 Similarly 16S-23S rDNA spacer-derived primers were designed by researchers that specifically amplified a 470 bp band in all strains of Xoo; as well as in other three Xanthomonas, including Xcc.34
In the present study, no amplification was observed with the Xoo-specific primer pair Xoo.txt+Xoo.txt4R in any samples, either from Xcp or Xcc (Table 4). Similarly, the infected pomegranate DNA samples did not yield any amplification.
In this study, we have developed an XRIC box-based PCR assay for the efficient diagnosis of Xcp infected samples. The SCAR primers designed from the XRIC box sequence were highly efficient in distinguishing between closely related Xanthomonas species. The present investigation showed close sequence similarity as well as cross-amplification between Xcp and Xcc. This analysis demonstrates the significance of the XRIC sequence in differentiation between Xanthomonas pathovars and specific diagnosis of Xcp. Currently, this assay is being used for diagnosis and allows for adapting appropriate management measures.
PCR amplification using the XRIC primer yielded two prominent bands of 216 bp and 159 bp in all Xcp and Xcc isolates as well as plant samples, including crude extracts. XRIC sequence study revealed that the initial 170 bases were highly conserved for Xcp, Xcc, Xcg, and Xcm. Therefore, the initial 170 bp XRIC region was a good candidate region for designing Xcp-specific primers. SCAR primer pair (Xcp1-20+Xcp133-152) thus designed amplified a 152 bp band in all Xcp isolates and infected pomegranate samples, except one isolate; while both Xcc isolates yielded 371 bp bands. Another SCAR primer pair, XcpF/R amplified a 200 bp and in all bacterial blight-infected plant samples and seven of eight Xcp isolates. An Xcp isolate along with both Xcc isolates amplified a 350 bp band. Reference KM-gyrB gene primers amplified a 491 bp and in all Xcp isolates, a 375 bp band in both Xcc isolates. However, PCR amplification showed 491 bp in 12 of 20 plant samples. Thus, it could be concluded that primers XRIC, SCAR-based XcpF/R, Xcp1-20+Xcp133-152, gyrase gene-based, and Xcc-specific primers will be very useful in the identification or detection of Xcp in diseased plant samples of pomegranate from fields. Both SCAR-based primers were designed in the present investigation to identify the bacterial pathogen Xcp without isolation of pomegranate bacterial blight culture. Therefore, it can be employed for efficient monitoring of the pathogen. These PCR primers identify the plant pathogen, i.e., bacteria directly from field samples. These results can also help to determine the best diagnostic and taxonomic position of the Xcp.
ACKNOWLEDGMENTS
The authors are thankful to the university authorities for providing the necessary facilities to undertake this work. The authors are also thankful to Mr. BA Hujare and Miss. SS Kadam for providing necessary isolates in the present study. Help provided by Dr. SG Borkar, Dr. AA Kale and Mr. BD Pawar during this study is also acknowledged.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Gaikwad GB, Nainwad RV, Borale SU and Gaikwad RB. Varieties of pomegranate (Punica granatum) in India. The Pharma Innovation Journal. 2023;12(12):2893-2901.
- Hingorani MK, Mehta PP. Bacterial leaf spot of pomegranate. Indian Phytopathol. 1953;5:55-56.
- Kamble SH, Surwase SN, Adagale VG, Khune SR, Bamankar SA, Mahadik PG. Evaluation of antimicrobial action of probiotics against Xanthomonas axonopodis pv. punicae causing bacterial blight of Punica granatum L. (Pomegranate). Annals of Plant and Soil Research. 2024;26(4): 633-642.
Crossref - Mondal KK and Sharma J. Bacterial blight: An emerging threat to pomegranate export. Indian Farming. 2009. 59(8):22-23.
- Sharma J, Sharma KK, Kumar A, et al. Pomegranate bacterial blight: symptomatology and rapid inoculation technique for Xanthomonas axonopodis pv. punicae. J Plant Pathol. 2017;99(1):109-119.
- Kumar P, Lokesh V, Doddaraju P, et al. Greenhouse and field experiments revealed that clove oil can effectively reduce bacterial blight and increase yield in pomegranate. Food Energy Secur. 2021;10(4):e305.
Crossref - Petersen Y, Mansvelt EL, Venter E, Langenhoven WE. Detection of Xanthomonas axonopodis pv. punicae causing a bacterial blight on pomegranate in South Africa. Australasian Plant Pathol. 2010;39(6):544-546.
Crossref - Icoz SM, Polat I, Sulu G, et al. First report of bacterial blight of pomegranate caused by Xanthomonas axonopodis pv. punicae in Turkey. Plant Dis. 2014;98(10):1427-1427.
Crossref - Catara V, Cubero J, Pothier JF, et al. Trends in molecular diagnosis and diversity studies for phytosanitary regulated Xanthomonas. Microorganisms. 2021;16:9(4):862.
- Hartung JS, Daniel JF, Pruvost OP. Detection of Xanthomonas campestris pv. citri by the polymerase chain reaction. Appl Environ Microbiol. 1993;59(4):1143-1148.
Crossref - Gillings MR, Fahy PC, Broadbent P, Barnes D. Rapid identification of a second outbreak of Asiatic citrus canker in the Northern Territory using the polymerase chain reaction and genomic fingerprinting. Australasian Plant Pathol. 1995;24(2):104-111.
Crossref - Hartung JS, Pruvost OP, Villemot I, Alvarez A. Rapid and sensitive colorimetric detection of Xanthomonas axonopodis pv. citri by immunocapture and nested-polymerase chain reaction assay. Phytopathology. 1996;86(1):95-101.
Crossref - Cubero J, Graham JH. Genetic Relationship among Worldwide Strains of Xanthomonas Causing Canker in Citrus Species and Design of New Primers for Their Identification by PCR. Appl Environ Microbiol. 2002;68(3):1257-1264.
Crossref - Pruvost O, Vital K, Ah-You N, Verniere C, Chiroleu F, Gagnevin L. A minisatellite-based MLVA for typing Xanthomonas citri pv. mangiferae indicae. J Plant Pathol. 2008;90(2):S2-362.
- Moretti C, Amatulli MT, Buonaurio R. PCR-based assay for the detection of Xanthomonas euvesicatoria causing pepper and tomato bacterial spot. Letters Appl Microbiol. 2009;49(4):466-471.
Crossref - Kale PB, Chimote VP, Raghuwanshi KS, Kale AA, Borkar SG. Microbial, biochemical, pathogenicity and molecular characterization of Xanthomonas axonopodis pv. punicae from pomegranate. J Pure Appl Microbiol. 2012;6(4):1699-1706.
- Giri MS, Prasanthi S, Kulkarni S, Benagi VI, Hegde YR. Biochemical and molecular variability among Xanthomonas axonopodis pv. punicae strains, the pathogen of pomegranate bacterial blight. Indian Phytopathol. 2011;64(1):1-4.
- Goel AK, Rajagopal L, Nagesh N, Sonti RV. Genetic locus encoding functions involved in biosynthesis and outer membrane localization of xanthomonadin in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2002;184(13):3539-3548.
Crossref - Raghuwanshi KS, Hujare BA, Chimote VP, Borkar SG. Characterization of Xanthomonas axonopodis pv. punicae isolates from western Maharashtra and their sensitivity to chemical treatments. Bioscan. 2013;8(3):845-850.
- Untergasser A, Cutcutache I, Koressaar T, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115-e115.
Crossref - San Millan RM, Martinez-Ballesteros I, Rementeria A, Garaizar J, Bikandi J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res Notes. 2013:6:513.
Crossref - Wang X, Liang S, Gan Q, Cai B,Liu C. Current status and future perspectives of the diagnostic of plant bacterial pathogens. Front Plant Sci. 2025;16:1547974.
Crossref - Hauben L, Vauterin L, Swings J, Moore ERB. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Intl J Syst Bacteriol. 1997;47(2):328-335.
Crossref - Mondal KK, Rajendran TP, Phaneendra C, et al. The reliable and rapid polymerase chain reaction (PCR) diagnosis for Xanthomonas axonopodis pv. punicae in pomegranate. African J Microbiol Res. 2012;6(30):5950-5956.
Crossref - Coletta Filho HD, Takita MA, De Souza AA, et al. Primers based on the rpf gene region provide improved detection of Xanthomonas axonopodis pv. citri in naturally and artificially infected citrus plants. J Appl Microbiol. 2006;100(2):279-285.
Crossref - Sakthivel N, Mortensen CN, Mathur S. Detection of Xanthomonas oryzae in artificially inoculated and naturally infected rice seeds and plants by molecular techniques. Appl Microbiol Biotechnol. 2001;56(3-4):435-441.
Crossref - Khan MI, Ur Rehma, M, Khan I, Shah TA, et al, Isolation, identification and characterization of Xanthomonas axonopodis pv. citri from selected species. Appl Ecol Environ Res. 2024:22(1):665-679.
Crossref - Bradbury JF. Xanthomonas Dowson (1939) In “Bergey’s Manual of Systematic Bacteriology”, Vol. 1 (eds N.R. Krieg and J.G. Holt), Williams & Wilkins, Baltimore, 1984:199-210.
- Sharma J, Sharma KK, Kumar A, et al. Pomegranate bacterial blight:symptomatology and rapid inoculation technique for Xanthomonas axonopodis pv. punicae. J Plant Pathol. 2017;99(1):109-119.
- Dutta A, Singh N. Genomic determinants for host adaptation or host specificity. In Microbial Genomics Volume 1. Host Adaptation, Virulence, and Evolution 2025;1:21-30.
Crossref - Johnson JS, Spakowicz DJ, Hong BY, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029.
Crossref - Sharma R, Sharma SD, Sharma VK, et al. First comprehensive evaluation of genetic variability for bacterial blight resistance in wild Punica granatum L. populations from the North-Western Himalayas. Physiol Mol Plant Pathol. 2024;134(17):102438.
Crossref - Chathalingath N, Gunasekar A, Venu S. Phenotypic and molecular characterisation of Xanthomonas axonopodis pv. punicae from pomegranate leaves. Physiol Mol Plant Pathol. 2023;128:102160.
Crossref - Adachi N, Oku T. PCR-mediated detection of Xanthomonas oryzae pv. oryzae by amplification of the 16S-23S rDNA spacer region sequence. J Plant Pathol. 2002;66(4):303-309.
Crossref - Vauterin L, Rademark J, Swings J. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathol. 2000;90(7):677-682.
Crossref - Parkinson N, Cowie C, Heeney J, Stead D. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol. 2009;59(2):264-274.
Crossref - Schaad NW, Postnikova E, Lacy G, Sechler A, et al. Emended classification of xanthomonad pathogens on citrus (Erratum). Syst Appl Microbiol. 2006;29(8):690-695.
Crossref - Mondal KK, Verma G, Mani C. Phylogenetic relatedness of Xanthomonas axonopodis pv. punicae, the causal agent of bacterial blight of pomegranate based on two loci, 16S rRNA and gyrB. Ann Microbiol. 2013;63:801-804.
Crossref - Radhika DH, Gunnaiah R, Lamani A, Peerjade D, Jagadeesha RC. Long read genome sequence resources of Xanthomonas citri pv punicae strain Bagalkot, causing pomegranate bacterial blight. Mol Plant-Microbe Interact. 2021;34(7):874-877.
Crossref - Yuan Z, Fang Y, Zhang T, et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol J. 2018;16(7):1363-1374.
Crossref - Doddaraju P, Kumar P, Gunnaiah R, et al. Reliable and early diagnosis of bacterial blight in pomegranate caused by Xanthomonas axonopodis pv. punicae using sensitive PCR techniques. Sci Rep. 2019;9(10097):1-9.
Crossref - Kumar P, Corrado G, Manjunatha G, et al. A Pseudomonas-based bio-formulation to control bacterial blight of pomegranate caused by Xanthomonas axonopodis pv. punicae. Biol Control. 2025;201:105686.
Crossref - Prasannakumar MK, Parivallal PB, Manjunatha C, et al. Loop-mediated isothermal amplification assay for pre-symptomatic stage detection of Xanthomonas axonopodis pv. punicae infection in pomegranate. Australasian Plant Pathol. 2020;49(3):467-473.
Crossref - Cubero J, Graham JH. The leucine-responsive regulatory protein (lrp) gene for characterization of the relationship among Xanthomonas species. Int J Syst Evol Microbiol. 2004;54(Pt 2):429-437.
Crossref - Brunings AM, Gabriel DW. Xanthomonas citri:breaking the surface. Mol Plant Pathol. 2003;4(3):141-157.
Crossref - Ngoc LB, Verniere C, Belasque JJ, et al. Ligation-mediated PCR, a fast and reliable technique for insertion sequence-based typing of Xanthomonas citri pv. citri. FEMS Microbiol Lett. 2008;288(1):33-39.
Crossref - Gupta VS, Rajebhosale MD, Sodhi M, et al. Assessment of genetic variability and strain identification of Xanthomonas oryzae pv. oryzae using RAPD-PCR and IS1112 based PCR. Curr Sci. 2001;80(8):1043-1049.
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.