ISSN: 0973-7510
E-ISSN: 2581-690X
Thattekad bird sanctuary, located in the Western Ghats of Kerala, India, which hosts an unexplored microbial community, is selected for the present investigation. Microbes play a major role in mineral recycling and nutrient absorption by the flora and fauna in the habitat. Various bacterial extracellular enzymes facilitate all these activities. The increasing demand for microbial enzymes in favor of green technology encouraged us to focus on exoenzyme profiling of bacterial isolates from forest soil samples. The present study is aimed at the screening and identification of exoenzyme producing soil bacterial strains isolated from evergreen forests and moist deciduous forests of Thattekad bird sanctuary. In this study, only multienzyme producing bacteria were selected for detailed analysis because such bacteria are highly relevant in multi-enzyme dependent processes such as biowaste degradation. We screened for nine hydrolytic exoenzymes namely, amylase, cellulase, ligninase, pectinase, xylanase, caseinase, gelatinase, esterase and lipase, and identified 79 multienzyme-producing bacterial strains, mostly belonging to phylum Firmicutes and Proteobacteria. Firmicutes from evergreen forests and moist deciduous forests produced a greater number of enzymes compared to Proteobacteria. Also, bacterial strains isolated from evergreen forest soil produced more enzymes compared to moist deciduous forest. Bacillus amyloliquefaciens strain TBS040 isolated from moist deciduous forest soil was found to produce all the nine enzymes screened. Enzymatic hydrolysis of biowaste using cell free crude enzyme extract from Bacillus velezensis strain TBS064 resulted in enhanced bioethanol production. These findings highlight the importance of screening unexplored habitats for the identification of novel strains, which can contribute to the future of green technology.
Forest Soil, Bacterial Exoenzyme Profiling, Functional Metagenomics, Bioethanol, Biowaste
Thattekad bird sanctuary in the Western Ghats is one of the richest avifauna habitats in peninsular India. It comprises diverse ecosystems ranging from tropical evergreen forests to riparian forests with a variety of flora and fauna. Based on the vegetation, the forest is divided into evergreen forests (EGF) and moist deciduous forests (MDF), where nearly 60% of the total area is the moist deciduous forest. Evergreen forests are further divided into tropical wet and semi-evergreen forests, which span about 20% of the total area. The remaining area of the sanctuary is comprised of riparian forests and plantations. It has been widely accepted that in forest ecosystems, the nature of vegetation and the affluence of nutrients in the soil are influenced by the microbial community.1-3
Species richness and endemism of flora and fauna in the Western Ghats have been studied extensively. However, very few attempts have been made to unravel the diversity of microorganisms in this area.4 Microorganisms are generally claimed to be the potential sources of bioactive molecules widely used in medical and industrial fields. Numerous soil microbes are integral components of biogeochemical cycles and decomposition of organic matter, which involve enzymatic reactions.
Because of potential biotechnological applications, the enzyme producing microbes have always been a major focus of research especially using soil as the microbial resource.5 Enzymes of microbial origin are widely applied in industries due to their availability, eco-friendliness, and cost-effective production options.6 Moreover, with the advancement of genomics, proteomics, and DNA recombinant methodologies, a microbe can be converted into a programmable chassis, balancing the needs and constraints during large-scale production.5 Bacterial enzymes are preferred for industrial applications over the fungal enzymes because of the fast rate of growth of bacteria and their compatibility to bioengineering processes. Multienzyme producing bacteria are of great advantage to industries which require the utilization of different enzymes at multiple levels during the production of a single product.6
Hence the present study was designed to isolate and identify the soil bacterial strains present in evergreen and moist deciduous forests of Thattekad Bird Sanctuary using culture-based methods and metagenomic analysis and also to profile the multienzyme producing strains. This study focuses on bacteria which secrete multiple hydrolytic enzymes such as amylase, cellulase, ligninase, pectinase, xylanase, caseinase, gelatinase, esterase and lipase. In order to validate the application of these exoenzymes, a study on the biofuel production from waste materials treated with an exoenzyme produced by the isolated bacterial strains was also carried out.
Sample collection
The soil was collected from different locations of Thattekad bird sanctuary that falls between 10°7′ and 11°N latitude, 76°40′ and 76°45′ E longitude. The soil samples were collected from each plot in sterilized container. EGF and MDF areas were divided into 10-12 square plots and 30-35 square plots of 500 X 500 M and the soil samples collected from each of these plots were used to isolate bacteria. For metagenomic analysis, all the samples from different vegetations were thoroughly mixed to form one composite sample under sterile conditions.
Isolation of culturable bacteria
The serial dilution technique was utilized to isolate culturable bacteria from soil. After serial dilution (10-1 to 10-7), 1 ml of the suspension was inoculated into nutrient agar using the spread plate technique. Following inoculation, the plates were kept in the incubator at 25°C temperature for up to 3 days, and colonies of different morphologies were selected for screening. Stock cultures were maintained as glycerol stock at −80°C and stored for further studies.7
Screening and profiling of extracellular enzymes
All the isolates were subjected to optimum conditions to facilitate the production of the following enzymes: amylase, cellulase, ligninase, pectinase, xylanase, gelatinase, caseinase, esterase, and lipase. Most of the chemicals and biochemicals were purchased from HiMedia, India, except those which are mentioned separately. In order to screen the bacteria for enzyme production, isolates were initially grown in nutrient broth for 24 h at 28°C. One loopful was spotted on specific minimal media supplemented with an appropriate substrate and incubated at 28°C for detection of extracellular hydrolytic enzyme.8 Diameter of the colony and that of the halo zone was measured. The Enzymatic Index (EI) in each enzyme assay was measured after incubation, expressed as9 EI= (colony diameter + halo zone diameter)/colony diameter.
Amylase activity was determined by inoculating the strains in starch agar, and after incubation, the culture plates were flooded with 2% iodine solution for visualization of the halo zone around the positive colony.10 Cellulase activity was detected using carboxymethyl cellulose (CMC), as the substrate at 0.5% (w/v). After the incubation period, 0.2% congo red was poured into the plates kept for 15-30 min and then washed with 1 M NaCl (Sigma Aldrich, USA). The de-staining allowed the visualization of clear halos.11 The medium containing congo red (Sigma Aldrich, USA) was used for screening ligninase activity, and the decolorization zone around the microbial colonies indicated ligninase production.12 A minimal agar medium containing 0.5% oat spelt xylan, as the only carbon source, was used for the screening of xylanase activity. After incubation, 0.4% congo red dye was added to the petri plates and washed with 1M NaCl. The colonies which showed a clear halo was selected as the positive strains.13 Inoculation of strains in pectin agar was performed for pectinase producing bacteria and after the incubation period, the plates were observed for the formation of a clear zone around the colonies by flooding them with iodine solution.14
Proteolytic activity was screened using gelatin as substrate for gelatinase and skim milk media for caseinase. Clear zones around the colonies indicated that microbes are able to hydrolyze the protein.11,15 Screening in Tween 20 assessed esterase activity, and a white precipitate around the colonies indicated the presence of the enzyme. Tributyrin agar was used for the detection of lipase, and a zone of clearance surrounding the colony indicated a positive result.16
Statistical analysis
Qualitative enzyme analysis was carried out as six independent experiments. Statistical analyses were performed using one-way ANOVA followed by Post Hoc analysis by Tukey’s HSD test using SPSS version 12.
Identification of bacterial strains
The strains that produced the aforementioned enzymes were selected for identification. The genomic DNA was extracted from bacterial strains using the XpressDNA Bacterial Genomic DNA kit (MagGenome Technologies, Chennai) in accordance with the manufacturer’s instructions. Universal bacterial primers were used to amplify 16S rDNA from the genomic DNA. Amplification was performed in T100 Thermocycler (Bio-Rad, USA). The conditions used for PCR amplification were: (a) initial denaturation at 95°C for 2 min, (b) 30 cycles of 95°C for 1 min, 58°C for 30s, 72°C for 1 min and (c) final extension at 72°C for 7 min. The 16S rDNA primers (Sigma-Aldrich) were 27F-5-AGAGTTTGATCCTGGCTCAG-3 (forward) and 1525R-5-TACGGYTACCTTGTTACGACTT-3 (reverse). The PCR products were sequenced by Sanger’s method (AgriGenome Labs Pvt. Ltd, Kochi). For the identification of bacteria, the sequences of 16S rRNA gene were analyzed using the BLAST-N search program at the National Centre for Biotechnology Information (NCBI) site. All of the bacterial sequences were submitted to NCBI GenBank for granting of accession numbers.
Functional diversity analysis
DNA isolation
For DNA extraction from soil samples DNeasy PowerSoil Kit (Qiagen) was used and was performed by following the manufacturer’s protocols. Quantitation of extracted DNA was done by a Qubit fluorometric analyzer (Thermo Fisher Scientific) and quality of the extracted DNA was checked by 0.8% agarose gel electrophoresis.
Library preparation
In order to analyze the microbiome community, metagenomic library preparation was performed in accordance with manufacturer’s instructions of KAPA Hyper Prep Kit by ROCHE. Paired end libraries were constructed with the insert size of 250 and 350 bp from 250 µg of genomic DNA as sample DNA input.
Sequencing and metagenomic analysis
High-throughput sequencing was carried out on HiSeq 2500 system (Illumina) using paired-end reads of length 2 × 150 bp (Bionivid Technology, Bangalore). Samples were sequenced using 150 bp paired-end module sequencing. For quality checking, raw read was subjected to quality control analysis using fastp tool to obtain high quality (HQ) filtered reads. The sample was assembled using metaspades assembler individually with default parameters where k-mer length chosen is 91. The statistical elements of the assemblies were calculated using NGSQC toolkit. MetaErg pipeline was used on default parameters to evaluate the functional abilities of microbial communities present in the samples.
Bioprospection of cellulase enzyme for bioethanol production
Enzymatic Assay of cellulolytic activity
Enzymatic Assay of cellulolytic activity of bacterial strains was performed according to the method of Bailey et al.,17 with slight modifications. 0.5 mL of 1% (w/v) carboxymethyl cellulose (CMC) prepared in 50 mM phosphate buffer (pH 7) was added into test tubes, to which 0.5 ml of the cell free extract from the production medium of different bacterial cultures were added. This was followed by the incubation of enzyme-substrate mixture at 50°C for 10 min. In order to terminate the reaction, 1.5 ml of 3, 5-dinitrosalicylic acid (DNS), Sigma Aldrich, USA) reagent was added. Enzyme blanks and controls containing all of the reagents were run in parallel. In the enzyme blank, the reaction was terminated before the addition of cell free extract. In the control sample, reaction was stopped and distilled water was added instead of cell free extract. The tubes were placed in boiling water for 10 minutes and cooled down to room temperature in water to ensure stabilization. The amounts of reducing sugars thus released were measured at 540 nm in the spectrophotometer (Shimadzu UV-1800 Japan), using glucose as the standard for reducing sugar. One unit of cellulase is defined as the amount of enzyme that liberates 1 μmol of glucose equivalents per minute /mL of culture supernatant under the standard assay conditions.
Estimation of cellulose
Two biowaste materials, namely, banana peels and pineapple leaves, were collected, cut into small pieces, oven-dried and powdered. Then the total cellulose content in both samples was determined by the Anthrone method.18
Enzymatic hydrolysis
The best cellulase producing strain TBS064 along with standard strain from NCIM, Cellulomonas uda NCIM 2353/5351 was used for the pretreatment of samples for hydrolyzing cellulose to liberate reducing sugar for fermentation. These bacterial strains were inoculated in CMC broth media to facilitate the production of cellulase. After 48 hrs of incubation, the production media was centrifuged (Eppendorf Centrifuge 5418, Germany); the supernatant was collected and used as the source of enzyme. Biowaste samples were sterilized by autoclaving at 120°C for 15 min. 10 g of the sterilized samples were treated with 10 ml of culture supernatant in 90 ml of phosphate buffer (pH 7). Enzymatic hydrolysis was performed at 30°C in a 250 ml stoppered flask with gentle agitation at 100 rpm and incubated for 48 hours. The samples were centrifuged at 10000 rpm for 5 min at 4°C and the supernatant was collected and filter sterilized. This hydrolysate was then used for bioethanol production.
Bioethanol production
Inoculum of Saccharomyces cerevisiae strain was prepared in 10.0 ml of Yeast Peptone Dextrose medium at pH 5.5 and incubated at 25°C for 48 hrs. The yeast strain was acclimatized by a series of successive liquid cultures with increasing amounts of pretreated hydrolysate of biowaste. Fermentation was carried out using the enzyme treated biowaste hydrolysate from both the substrates along with untreated controls replacing malt in the fermentation media. During the process, the fermented product was collected at regular intervals of 24, 48, and 72 hrs, centrifuged at 7000 rpm for 10 min to collect the supernatant. Ethanol concentration was determined by dichromate method using a microplate reader (iMark™ Microplate Absorbance Reader- Bio-Rad, USA) after phase separation by tri-n-butyl-phosphate,19 (Sigma Aldrich, USA).
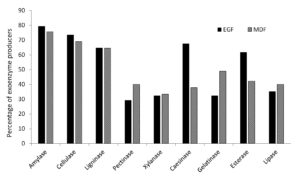
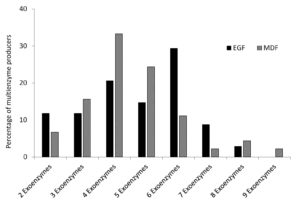
A total of 59 and 77 soil bacterial strains were isolated from EGF and MDF of Thattekad bird sanctuary, respectively. Subsequently, 79 strains (34 from EGF samples and 45 from MDF samples), which produced two or more enzymes on repeated sub-culturing and screening, were selected for detailed analysis. During the study, we analyzed the production of nine exoenzymes and it was observed that the percentage of exoenzyme producers among soil bacteria in EGF and MDF samples are comparable except for caseinase and esterase, which are high in EGF and gelatinase, which is slightly higher in MDF samples (Figure 1). Among the 34 strains isolated from EGF soil samples, 79.41% produced amylase and 73.53% produced cellulase. It was also found that ligninase was produced by 64.71% of bacterial strains, pectinase by 29.41%, xylanase by 32.35%, caseinase by 67.65%, gelatinase by 32.35%, esterase by 61.76% and lipase by 35.29%. In MDF soil samples, among the 45 strains the percentages are 75.56%, 68.89%, 64.44%, 40%, 33.33%, 37.78%, 48.89%, 42.22% and 40% respectively for producers of amylase, cellulase, ligninase, pectinase, xylanase, caseinase, gelatinase, esterase and lipase (Figure 1). Bacterial strains isolated from EGF produced more enzymes compared to MDF samples. In EGF samples, 41.1% of the soil bacterial strains produced 6 or more exoenzymes while 58.9% produced 5 or less. For MDF it is 20% and 80%, respectively (Table S1, Table S2 and Figure 2).
Soil bacterial strains screened for nine exoenzymes are shown in the X-axis, and the percentage of bacterial strains, which produce each of those enzymes, is shown in the Y-axis. Percentage of exoenzyme producers from among 34 soil bacterial strains of Evergreen Forest (EGF) and 45 bacterial strains of Moist Deciduous Forest (MDF) from Thattekad bird Sanctuary are compared and represented in the figure.
Figure 1. Percentage of exoenzyme producing soil bacteria from EGF and MDF
Exoenzymes, produced by bacterial strains isolated from EGF and MDF. Number of enzymes produced by the bacterial strains are shown in the X axis and percentage of bacterial strains which produced multienzymes are shown in Y axis.
Figure 2. Percentage of multiple exoenzyme producing strains from EGF and MDF
These 79 bacterial strains were identified by molecular characterization using 16S rDNA sequencing and the sequences were deposited in NCBI GenBank. The names and corresponding accession numbers generated from NCBI for strains isolated from EGF and MDF soil samples are shown in Table 1 and Table 2, respectively.
Table (1):
Identity and Accession numbers of soil bacteria isolated from EGF samples.
No. |
Identity of the isolated bacterial strains |
Accession Numbers |
|---|---|---|
1 |
Pseudomonas aeruginosa strain TBS001 |
MW321482 |
2 |
Bacillus sp. strain TBS002 |
MW362562 |
3 |
Pantoea sp. strain TBS003 |
MW418329 |
4 |
Bacillus subtilis strain TBS004 |
MW418330 |
5 |
Bacillus toyonensis strain TBS005 |
MW418331 |
6 |
Bacillus cereus strain TBS006 |
MW418332 |
7 |
Bacillus cereus strain TBS007 |
MW418333 |
8 |
Paenibacillus alvei strain TBS008 |
MW418334 |
9 |
Bacillus albus strain TBS009 |
MW418335 |
10 |
Bacillus cereus strain TBS010 |
MW418336 |
11 |
Acinetobacter rhizosphaerae strain TBS011 |
MW418337 |
12 |
Lysinibacillus macroides strain TBS012 |
MW418338 |
13 |
Enterobacter cloacae strain TBS013 |
MW418339 |
14 |
Acinetobacter sp. strain TBS014 |
MW418340 |
15 |
Enterobacter hormaechei subsp. Xiangfangensis strain TBS015 |
MW418341 |
16 |
Atlantibacter hermannii strain TBS016 |
MW418342 |
17 |
Bacillus sp. strain TBS017 |
MW418343 |
18 |
Bacillus cereus strain TBS018 |
MW418344 |
19 |
Enterobacter sp. strain TBS019 |
MW418345 |
20 |
Bacillus sp. strain TBS020 |
MW418346 |
21 |
Enterobacter sp. strain TBS021 |
MW418347 |
22 |
Pseudomonas sp. strain TBS022 |
MW418348 |
23 |
Bacillus sp. strain TBS023 |
MW418349 |
24 |
Enterobacter cloacae strain TBS024 |
MW418350 |
25 |
Serratia sp. strain TBS025 |
MW418351 |
26 |
Pseudomonas monteilii strain TBS026 |
MW418352 |
27 |
Stenotrophomonas maltophilia strain TBS027 |
MW418353 |
28 |
Brevibacillus parabrevis strainTBS028 |
MW418354 |
29 |
Lysinibacillus xylanilyticus strainTBS029 |
MW418355 |
30 |
Bacillus tequilensis strain TBS 030 |
MW418356 |
31 |
Lysinibacillus xylanilyticus strain TBS031 |
MW418357 |
32 |
Lysinibacillus macroides strain TBS032 |
MW418358 |
33 |
Brevibacillus parabrevis strain TBS033 |
MW418359 |
34 |
Bacillus subtilis strain TBS034 |
MW418360 |
34 Bacterial strains were isolated from EGF soil samples after screening for 9 exoenzymes and were identified by 16S rDNA sequencing. Details of 34 organisms from EGF along with the accession number from National Center for Biotechnology Information (NCBI) is given in the table.
Table (2):
Identity and Accession numbers of soil bacteria isolated from MDF samples.
No. |
Identity of the isolated bacterial strains |
Accession Numbers |
|---|---|---|
1 |
Aeromonas veronii strain TBS035 |
OM900117 |
2 |
Enterococcus sp. strain TBS036 |
OM900118 |
3 |
Bacillus rugosus strain TBS037 |
OM900119 |
4 |
Pseudomonas sp. strain TBS038 |
OM900120 |
5 |
Ralstonia sp. strain TBS039 |
OM900121 |
6 |
Bacillus amyloliquefaciens strain TBS040 |
OL823001 |
7 |
Bacillus thuringiensis strain TBS041 |
OM900122 |
8 |
Bacillus cereus strain TBS042 |
OM900123 |
9 |
Aeromonas hydrophila strain TBS043 |
OM900124 |
10 |
Bacillus infantis strain TBS044 |
OM960966 |
11 |
Brevibacillus borstelensis strain TBS045 |
OM900125 |
12 |
Priestia aryabhattai strain TBS046 |
OM900126 |
13 |
Pseudomonas fluorescens strain TBS047 |
OM900127 |
14 |
Priestia megaterium strain TBS048 |
OM900128 |
15 |
Pseudomonas aeruginosa strain TBS049 |
OM900129 |
16 |
Bacillus tropicus strain TBS050 |
OL790352 |
17 |
Enterococcus faecalis strain TBS051 |
OM900130 |
18 |
Klebsiella sp. strain TBS052 |
OM900131 |
19 |
Priestia aryabhattai strain TBS053 |
OM900132 |
20 |
Brevibacillus borstelensis strain TBS054 |
OL790351 |
21 |
Bacillus pumilus strain TBS055 |
OM900133 |
22 |
Kurthia gibsonii strain TBS056 |
OM900134 |
23 |
Paenalcaligenes suwonensis strain TBS057 |
OM900135 |
24 |
Escherichia coli strain TBS058 |
OM900136 |
25 |
Pseudomonas lactis strain TBS059 |
OM960965 |
26 |
Pseudomonas fluorescens strain TBS060 |
OM900137 |
27 |
Escherichia coli strain TBS061 |
OM900138 |
28 |
Staphylococcus sp. strain TBS062 |
OM960964 |
29 |
Staphylococcus hominis strain TBS063 |
OM967172 |
30 |
Bacillus velezensis strain TBS064 |
OL790345 |
31 |
Lysinibacillus sp. strain TBS065 |
OM900139 |
32 |
Bacillus tequilensis strain TBS066 |
OM900140 |
33 |
Staphylococcus arlettae strain TBS067 |
OM900141 |
34 |
Bacillus proteolyticus strain TBS068 |
OM900142 |
35 |
Bacillus subtilis strain TBS069 |
OL790348 |
36 |
Ralstonia solanacearum strain TBS070 |
OL790347 |
37 |
Pseudomonas sp. strain TBS071 |
OM900143 |
38 |
Lysinibacillus macroides strain TBS072 |
OL790349 |
39 |
Lysinibacillus sphaericus strain TBS073 |
OM900144 |
40 |
Enterobacter sp. strain TBS074 |
OM900145 |
41 |
Microbacterium sp. strain TBS075 |
OM900146 |
42 |
Stenotrophomonas maltophilia strain TBS076 |
OM900147 |
43 |
Pseudomonas canadensis strain TBS077 |
OL790350 |
44 |
Niallia circulans strain TBS078 |
OM967175 |
45 |
Citrobacter portucalensis strain TBS079 |
OM967163 |
45 bacterial strains were isolated from MDF soil samples after screening for 9 exoenzymes and were identified by 16S rDNA sequencing. Details of 45 organisms from MDF along with the accession number from National Center for Biotechnology Information (NCBI) is shown in the table.
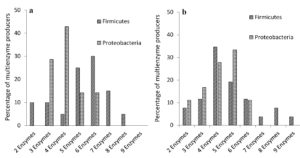
Among the multienzyme producers from soil samples collected from both these sites, organisms belonging to phylum Firmicutes and Proteobacteria were predominant. Details of multienzymes produced by Firmicutes and Proteobacteria were compared and represented in Figure 3. We directly compared the percentages of organisms, which produced more than one enzyme (2 to 9 enzymes) in both groups, Firmicutes and Proteobacteria. Among them, Firmicutes from both habitats produced greater number of enzymes compared to Proteobacteria, which probably makes them more flexible to substrate usage. In EGF, 50% of Firmicutes produced 6 or more exoenzymes, and remaining 50% produced 5 or less exoenzymes. In the case of Proteobacteria, the respective percentages are 14.29% and 85.72%. In MDF, 26.93% of Firmicutes produced 6 or more exoenzymes, whereas 73.08% produced 5 or less exoenzymes. For Proteobacteria the respective percentages are 11.11% and 88.89%.
Percentage of multienzymes produced are represented. (a) Firmicutes & Proteobacteria in EGF. (b) Firmicutes & Proteobacteria in MDF
Figure 3. Percentage of multiple exoenzyme producing Firmicutes and Proteobacteria
In EGF samples, among the 34 strains isolated, 20 were Firmicutes and 14 were Proteobacteria. The Brevibacillus parabrevis strain TBS028, that produced 8 exoenzymes was identified as the best multienzyme producer among the isolates from EGF samples and it showed the highest Enzyme Index (EI) for cellulase and pectinase (Table 3). In MDF samples, among the 45 strains isolated, 26 were Firmicutes, 18 were Proteobacteria and one was Actinobacteria. B. amyloliquefaciens strain TBS040 isolated from MDF samples is found to be the best multienzyme producer among all the 79 strains isolated from both habitats. It was the only organism that produced all the 9 enzymes that were screened. The best enzyme producing strains from both the sites are represented in Table 3.
Table (3):
Best exoenzyme producers among soil bacteria from EGF and MDF samples.
| No. | Enzyme | Site | Strain | EI (cm) |
|---|---|---|---|---|
| 1 | Amylase | EGF | Bacillus subtilis strain TBS034 | 6.31±0.14 |
| MDF | Pseudomonas canadensis strain TBS077 | 6.87±0.06 | ||
| 2 | Cellulase | EGF | Brevibacillus parabrevis strain TBS028 | 6.83±0.14 |
| MDF | Bacillus velezensis strain TBS064 | 7.33±0.24 | ||
| 3 | Ligninase | EGF | Bacillus sp. strain TBS020 | 6.37±0.20 |
| MDF | Bacillus velezensis strain TBS064 | 6.68±0.54 | ||
| 4 | Pectinase | EGF | Brevibacillus parabrevis strain TBS028 | 6.18±0.14 |
| MDF | Brevibacillus borstelensis strain TBS054 | 6.47±0.30 | ||
| 5 | Xylanase | EGF | Lysinibacillus xylanilyticus strain TBS029 | 6.44±0.16 |
| MDF | Lysinibacillus macroides strain TBS072 | 5.40±0.21 | ||
| 6 | Caseinase | EGF | Lysinibacillus xylanilyticus strainTBS031 | 6.15±0.05 |
| MDF | Brevibacillus borstelensis strain TBS054 | 5.62±0.22 | ||
| 7 | Gelatinase | EGF | Bacillus cereus strain TBS006 | 6.77±0.27 |
| MDF | Bacillus tropicus strain TBS050 | 7.64±0.15 | ||
| 8 | Esterase | EGF | Bacillus sp. strain TBS017 | 5.44±0.11 |
| MDF | Bacillus amyloliquefaciens strain TBS040 | 5.51±0.25 | ||
| 9 | Lipase | EGF | Enterobacter sp. strain TBS021 | 5.00±0.07 |
| MDF | Bacillus amyloliquefaciens strain TBS040 | 5.59±0.29 |
Best enzyme producer from EGF and MDF for each enzyme is shown along with the EI value.
All 79 bacterial isolates were screened for nine hydrolase enzymes as described earlier, and the Enzyme Indices (EI) were calculated for each, in order to assess the efficiency of enzyme production. These studies were conducted as six independent experiments and the results are given as supplementary data (Table S1 and Table S2). Results obtained from these studies were subjected to statistical analysis such as descriptive statistics, one-way ANOVA and Post Hoc using Tukey HSD test. It was evident from the statistical analysis that there was a significant difference in mean zone of EI at 5% level of significance for all enzymes with a high F value, indicating the significance of the model. Post Hoc analyses indicate that the variability among different strains, which produce a particular exoenzyme, will be high if the number of homogeneous groups is more. The statistical analysis data for each enzyme, including the range of mean values, are provided in Table 4. For example, for amylase, the Post Hoc tests showed 8 homogeneous groups of 27 positive strains for EGF and 17 homogeneous groups of 34 strains for MDF. It indicates that variability among amylase producers is higher in MDF than those in EGF. Similar results for all other enzymes are given in Table 4. It is evident from these results that for amylase and caseinase, the strains from MDF showed higher variability compared to those from EGF. On the other hand, for pectinase, xylanase and lipase, the strains from EGF exhibited higher variability. For the other enzymes viz., cellulase, ligninase, gelatinase and esterase, the variability among different strains was found to be similar at both sites.
Table (4):
Post Hoc analysis by Tukey’s HSD test of exoenzyme producing bacterial strains from EGF and MDF.
| No. | Enzymes | Site | Homogeneous groups | Number of Positive strains | Range of mean values |
|---|---|---|---|---|---|
| 1 | Amylase | EGF | 8 | 27 | 2.2650 – 6.3117 |
| MDF | 17 | 34 | 2.5800 – 6.8733 | ||
| 2 | Cellulase | EGF | 10 | 25 | 2.5894 – 6.8300 |
| MDF | 12 | 31 | 2.1800 – 7.3333 | ||
| 3 | Ligninase | EGF | 11 | 23 | 2.0200 – 6.3717 |
| MDF | 12 | 29 | 2.5400 – 6.6767 | ||
| 4 | Pectinase | EGF | 7 | 11 | 2.1967 – 6.1817 |
| MDF | 7 | 29 | 2.6167 – 6.4733 | ||
| 5 | Xylanase | EGF | 9 | 11 | 3.9670 – 6.4350 |
| MDF | 3 | 16 | 2.5633 – 5.4583 | ||
| 6 | Caseinase | EGF | 4 | 23 | 3.3676 – 6.1742 |
| MDF | 9 | 18 | 0.9250 – 5.6183 | ||
| 7 | Gelatinase | EGF | 6 | 12 | 0.5900 – 6.7733 |
| MDF | 15 | 22 | 2.2567 – 7.5383 | ||
| 8 | Esterase | EGF | 9 | 21 | 3.2433 – 5.5353 |
| MDF | 9 | 19 | 2.3967 – 5.5067 | ||
| 9 | Lipase | EGF | 6 | 12 | 2.4533 – 4.9950 |
| MDF | 6 | 18 | 2.6383 – 5.5933 |
One-way ANOVA followed by Post Hoc Analysis by Tukey HSD was done to each exoenzyme producing strains on the basis of EI.
In order to gain an in-depth understanding of the enzyme production profiles of microbiome from these habitats, we pursued shotgun sequencing and metagenomic analysis. The metagenome annotation predicted 325 bacterial genes of which 109 were 16S rRNA, 189 were 23S rRNA and 27 were 5S rRNA. The raw metagenome sequence was submitted to the SRA division of Genbank database in NCBI with accession number PRJNA820576. Protein domain analysis was carried out using MetaErg program, the pipeline that utilizes searches like Hidden Markov Model, BLAST and Diamond for annotation and visualization of metagenomic contigs.
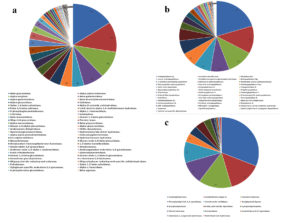
The annotated genes were searched against Swissport, Pfam, Tigrfam, FOAM, metabolic hmm, and genome diamond for protein domains and results of three biomolecule hydrolases are sorted and given in the figure. (a)Carbohydrate Hydrolases, (b) Proteolytic Hydrolases, (c) Lipolytic Hydrolases.
Figure 4. Hydrolases identified by metagenomic functional analysis
Since the initial focus of our investigation was on nine hydrolytic exoenzymes, subsequent functional metagenomic analyses were restricted to hydrolase enzymes. Hydrolases specific for carbohydrate, protein and lipids were sorted from the pool of data. The analysis revealed the presence of 54 different carbohydrate hydrolases, 40 protein hydrolases and 14 lipid hydrolases. It was observed that among the hydrolases, carbohydrate hydrolases were most abundant, followed by protein hydrolases and lipid hydrolases (Figure 4). The predominant carbohydrate hydrolases included polysaccharide hydrolases like betaglucosidase, alpha – alpha trehalase, alpha amylase, beta and alpha galactosidase, alpha glucosidases, cellulases, and alpha, beta xylosidases. Chitinases, amylases, trehalases, pullulanases, rhamnosidases, dextrinases, sucrases, fucosidases and agarases were also present. Major protein hydrolases were endopeptidases, peptidases, cysteine sulfatases, aminopeptidases like leucyl aminopeptidases, methionyl aminopeptidases, beta peptidyl aminopeptidases, Xaa-pro aminopeptidases, carboxyl peptidases and oligopeptidases. Prepilin peptidase, microbial collagenases and separases were also detected. Lipid hydrolases include carboxyl- esterases, arylsulfatases, cocaine esterases, acylglycerol lipase, epoxidases, sterol esterases, ceramidases and palmitoyl – CoA hydrolases.
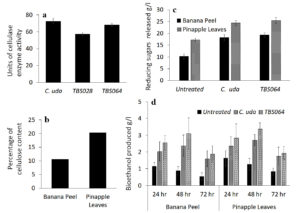
In order to demonstrate the functional application of a representative exoenzyme, we assessed bioethanol production from biowaste materials, which were treated with cellulase produced by two bacterial strains. Cellulolytic activity in the culture supernatant was assessed quantitatively for the strains TBS028 and TBS064 in comparison with the standard strain Cellulomonas uda, NCIM 2353/5351 and the results are given in Figure 5a. Cellulomonas uda showed the highest activity having 72.6U followed by strain TBS064 with 68.1U and strain TBS028 with 57.0U. Strain TBS064 was selected for further studies.
The cellulose content of two locally available biowaste materials, namely, banana peels and pineapple leaves were determined and the results are shown in Figure 5b. Though pineapple leaves contained more cellulose (20.34%) than the banana peel (10.63%), both these biowaste materials were used as substrates for bioethanol production. The cellulolytic efficiency of cell free crude enzyme prepared from the strain TBS064 was tested on both these biowastes. Slurry prepared with the biowaste in phosphate buffer was used as the substrate and incubated with cell-free crude enzyme extract from strain TBS064 and Cellulomonas uda independently. The amount of total reducing sugars liberated from banana peel and pineapple leaves after 48hrs of enzymatic hydrolysis was determined by DNS method. The amount of reducing sugars produced from pineapple leaves treated with cellulase enzyme from strain TBS064 (25.43g/l) was the highest among all (Figure 5c). Similar results were obtained for banana peels as well, where the sugar content was 19.28g/l and 18.30g/l for B. velezensis strain TBS064 and Cellulomonas uda, respectively. The bioethanol production from slurry filtrate of biowaste (enzyme treated and untreated) fermented in the presence of Saccharomyces cerevisiae was estimated periodically for 24 to 72 hrs (Figure 5d). Maximum production of bioethanol was observed after 48 hrs of incubation of both pineapple leaves and banana peel hydrolysate treated with cell free extract from strain TBS064 (3.38g/l and 3.1g/l). Bioethanol production from enzyme treated samples was found to be 1.7-1.8 times higher than that of untreated samples, which indicates that the soil bacterial B. velezensis strain TBS064 is a promising cellulase producer with potential industrial applications.
Estimation of cellulase enzyme production and analysis of bioethanol production by the potent isolates. (a) Exoenzyme production in the culture supernatant of TBS028, strain TBS064 in comparison with a standard strain Cellulomonas uda, NCIM 2353/5351. (b) The cellulose content of the locally available biowastes banana peel and pineapple leaves.(c) The total sugar liberated from banana peel and pineapple leaves after 48hrs of enzymatic hydrolysis with TBS064 and Cellulomonas uda 2353/5351. (d) The bioethanol production of enzyme treated and untreated slurry filtrate of biowaste fermented with Saccharomyces cerevisiae for 24-72 hrs
Figure 5. Assessment of bioethanol production from biowaste treated with cellulase from strain TBS064 for bioprospection
Western Ghats recognized as the global biodiversity hot spot, has prolific ecosystems and it could be a dynamic source of functionally active microbial community.20 This conserved ecosystem may contain unidentified microbes and the soil of the Western Ghats is often explored for industrially important metabolite producing microbes.21 Soil bacteria can be exploited for their ability to produce different enzymes of industrial significance. Enzymes of microbial origin should be promoted for industrial applications in support of the concept of green chemistry, wherein the use of eco-friendly and renewable raw materials is promoted for the synthesis of commercial products.22 In this context, we decided to study the soil bacterial strains of the evergreen forests and moist deciduous forests of Thattekad bird sanctuary, a microbiologically unexplored area in the Western Ghats. In the present study, we report the exoenzyme profiling of 79 bacterial strains, which have the capacity of multienzyme production, and the effect of enzymes produced by one such bacterium on bioethanol production. Earlier studies on bacterial enzyme production have pointed out their uniqueness, versatility and performance in extreme conditions of temperature and pH during the downstream processes, thus promoting their usage in various bioprocesses.23 Isolation of multienzyme producing bacteria is of great significance in various commercial applications since they are able to make biochemical processes faster and cost effective.6, 24
In our study amylase was found to be the enzyme produced by a maximum number of strains from both sites. We isolated 27 amylase producers from EGF and 34 from MDF which together account for 79% of all isolated strains. Amylase, the enzyme used specifically for starch degradation has been widely used in several industries like food, medicine, pharmaceuticals, etc.25 56 cellulase-producing strains have been isolated from these two sites, which form 73% of all the strains. Cellulase is recognized as a major biocatalyst in various industries because of the wide use of cellulose substrates.26 Ligninase producers were also present significantly among these strains and 52 ligninase producers (67.5%) were isolated in this study. Extracellular ligninolytic bacteria are gaining popularity as a source of enzymes in the biofuel industry and are very relevant in this new era of green biotechnology.27 Also, 27 xylanase producers (35%) and 28 pectinase producers (36%) were isolated from these sites. Xylanase and pectinase producers were few in number compared to all other enzyme producers. Both these enzymes have a multitude of uses in various industries particularly in the manufacture of food products.28 In addition, 40 caseinase producers (52%) and 33 gelatinase producing strains (43%) were isolated from these habitats. Protease enzymes, especially caseinase and gelatinase of bacterial origin, play pivotal roles in research and pharmaceutical areas.29 Screening for lipolytic organisms showed that esterases were produced by more strains than lipases. These enzymes have got tremendous applications in various industries ranging from the manufacture of detergents to medicines.30 It is anticipated that novel biocatalysts from microbes can offer a solution for the growing demand for enzymes in industries. These findings highlight the importance of screening unique ecosystems for the identification of novel strains. This warrants further research in understanding the unique features, if any, of these enzymes and to make them accessible for industrial applications.
It is evident that the predominant enzyme producers in these habitats belonged to phylum Firmicutes. The presence of several types of enzymes in higher quantities in Firmicutes clearly implies their active metabolic potential. It was also reported previously that Firmicutes are considered as metabolically versatile with various types of multienzyme producers.31 These bacteria are a boon to several industries that require the performance of different enzymes at multiple levels during the production of a single product.6 Among the Firmicutes, the predominant strains belonged to the genus Bacillus. Bacillus is a major bacterial genus found in the soil with various ecological functions, and because of their adaptability, they are valuable to various industries.32 Another 32 exoenzyme producing strains belonging to phylum Proteobacteria and one strain belonging to Actinobacteria were also isolated. The dominance of Firmicutes and Proteobacteria that produce extracellular enzymes has been reported in soil microbial diversity studies.33 The ability of Bacillus species to produce a broad spectrum of metabolites has been reported previously.34
Details of hydrolases present in the soil bacterial strains were also collected from the functional diversity data procured through metagenomic analysis. Screening of enzymes using the metagenomic approach is a quicker way to resolve the need of novel efficient biocatalysts for industrial applications. There are various reports on patented enzymes derived using this approach, which are currently used in various industries.35 In metagenomics, the functional analysis reveals all metabolic pathways of microbes in an ecosystem and the metabolites produced by them. This information has become very useful for the identification of novel and stable enzymes with economic significance for industrial purposes.36 In this study, we have used MetaErg pipeline for functional analysis, which is a robust open-source platform for metagenome analysis. It is useful for gene prediction and metabolic pathway analysis.37 Hydrolases are one of the best representative groups among enzymes used for various biotechnological applications38 and are the main focus in many metagenomics analyses.39 In our study, among the hydrolases, carbohydrate hydrolases were found to be more abundant compared to other hydrolases and 54 different carbohydrate hydrolases were present. All these enzymes belong to the glycoside hydrolases family, which helps the microbes in biomass utilization and have got a variety of industrial applications from pharmaceutical to cosmetic industries.40 The metagenomic analysis also showed the presence of 40 different protein hydrolases like endopeptidases, peptidases, cysteine sulfatases, and aminopeptidases. There are several proteases currently used in industries, which were created from soil metagenomic libraries after the functional analysis.41 Metagenomics analysis also identified 14 different lipid hydrolases including esterases like carboxyl- esterases, cocaine esterases and lipases like acylglycerol lipase that can be used in various food industries.42 Identification of the afore-mentioned hydrolase enzymes is a major outcome of the present investigation.
To demonstrate the functional application of a representative exoenzyme we performed the conversion of cheap biomass to biofuel using B. velezensis strain TBS064, which showed the highest EI for cellulase enzyme. Ethanol production using banana peel and pineapple leaves as the raw material was selected as a model system for evaluating the effect of the enzymatic treatment. Use of enzymes for the liberation of sugars from cellulose-rich biomass can be a viable strategy for eco-friendly and economical means of ethanol production. In the present study, we compared ethanol production from biowastes in the presence and absence of exoenzymes of bacterial origin from the B. velezensis strain TBS064. Analysis showed that the bioethanol production from enzyme treated biowaste substrate was at least 1.7 times higher than the untreated biomass. This result is a clear indication that a bacterial exoenzyme can play a key role in enhancing biofuel production and hence many of these enzymes hold the potential to improve current biochemical processes.
From this study, we identified 79 multienzyme producing bacterial strains mainly belonging to phylum Firmicutes and Proteobacteria. B. amyloliquefaciens strain TBS040 which produces nine extracellular enzymes can be a promising candidate for ecofriendly industrial applications. Bioethanol production from enzyme treated samples was found to be 1.7 times higher than that of untreated samples, showing the efficiency of the best cellulase producer, B. velezensis strain TBS064. Metagenomic analysis showed the presence of 54 carbohydrate hydrolases, 40 protein hydrolases and 14 lipid hydrolases. The results show that the selected area with rich biodiversity is a dynamic ecosystem, which harbour various microorganisms that can be explored for potential biotechnological applications.
Additional file: Additional Table S1 and S2.
ACKNOWLEDGMENTS
The authors would like to thank Chief Wildlife Warden, Kerala Forests and Wildlife Department, for the kind approval for site visit and sample collection.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AT and MS designed the work and wrote the manuscript. SR guided with the geographical data and sample collection. SPM helped us with the statistical analysis of the data. AT, MS, SR and SPM analyzed the data. All authors read and approved the manuscript for publication.
FUNDING
This study was funded by SARD scheme of Kerala State Council for Science Technology and Environment, Thiruvananthapuram, and the FIST scheme of the Department of Science and Technology, New Delhi, India.
DATA AVAILABILITY
DNA sequence data that support the findings of this research have been deposited in NCBI GenBank database and the accession code for the same is provided in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Wei H, Peng C, Yang B, et al. Contrasting Soil Bacterial Community, Diversity, and Function in Two Forests in China. Front Microbiol. 2018;9:1693.
Crossref - Liu L, Zhu K, Wurzburger N, Zhang J. Relationships between plant diversity and soil microbial diversity vary across taxonomic groups and spatial scales. Ecosphere. 2020;11(1):e02999.
Crossref - Fu Q, Shao Y, Wang S, et al. Soil Microbial Distribution Depends on Different Types of Landscape Vegetation in Temperate Urban Forest Ecosystems. Front Ecol Evol. 2022;10:858254.
Crossref - Rajeev AC, Sahu N, Deori M, Arun Dev S, Pal Yadav V, Ghosh I. Metagenomic Exploration of Microbial Signatures on Periyar River Sediments from the Periyar Tiger Reserve in the Western Ghats. Genome Announc. 2018;6(11):e00154-18.
Crossref - Choi JM, Han SS, Kim HS. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnology Advances. 2015;33(7):1443-1454.
Crossref - Kaur A, Rishi V, Soni SK, Rishi P. A novel multi-enzyme preparation produced from Aspergillus niger using biodegradable waste: a possible option to combat heterogeneous biofilms. AMB Expr. 2020;10(1):36.
Crossref - Lavanya J, Manjula S, Chitra AG, Chaithanya V, Sivaramakrishnan R. Efficacy of Peptone-glycerol Broth in Long-term Storage of the Bacterial and Yeast Cultures. JCDR. 2021;15(2):5-9.
Crossref - Charbonneau DM, Meddeb-Mouelhi F, Boissinot M, Sirois M, Beauregard M. Identification of Thermophilic Bacterial Strains Producing Thermotolerant Hydrolytic Enzymes from Manure Compost. Indian J Microbiol. 2012;52(1):41-47.
Crossref - Lechuga EGO, Zapata IQ, Nino KA. Detection of extracellular enzymatic activity in microorganisms isolated from waste vegetable oil contaminated soil using plate methodologies. Afr J Biotechnol. 2016;15(11):408-416.
Crossref - Lopez-Mondejar R, Zuhlke D, Becher D, Riedel K, Baldrian P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep. 2016;6(1):25279.
Crossref - Ghribi M, Meddeb-Mouelhi F, Beauregard M. Microbial diversity in various types of paper mill sludge: identification of enzyme activities with potential industrial applications. SpringerPlus. 2016;5(1):1492.
Crossref - Cui D, Li G, Zhao M, Han S. Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnology & Biotechnological Equipment. 2014;28(3):478-486.
Crossref - Shanthi V, Roymon M. Isolation, Identification and Partial Optimization of Novel Xylanolytic Bacterial Isolates from Bhilai-Durg Region, Chhattisgarh, India. Iran J Biotechnol. 2018;16(3):200-212.
Crossref - Shrestha S, Khatiwada JR, Zhang X, et al. Screening and Molecular Identification of Novel Pectinolytic Bacteria from Forest Soil. Fermentation. 2021;7(1):40.
Crossref - Balan SS, Nethaji R, Sankar S, Jayalakshmi S. Production of gelatinase enzyme from Bacillus spp isolated from the sediment sample of Porto Novo Coastal sites. Asian Pac J Trop Biomed. 2012;2(3):S1811-S1816.
Crossref - Ramnath L, Sithole B, Govinden R. Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol Rep. 2017;15:114-124.
Crossref - Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23(3):257-270.
Crossref - Updegraff DM. Semimicro determination of cellulose inbiological materials. Anal Biochem. 1969;32(3):420-424.
Crossref - Miah R, Siddiqa A, Tuli JF, et al. Inexpensive Procedure for Measurement of Ethanol: Application to Bioethanol Production Process. Adv Microbiol. 2017;07(11):743-748.
Crossref - Vasava ZT, Mistry KJ, Patel PP, Markande AR. Exploring microbial bioactive molecules from Western Ghats, India. Acta Ecologica Sinica. 2022;42(6):593-599.
Crossref - Siddharth S, Vittal RR, Wink J, Steinert M. Diversity and Bioactive Potential of Actinobacteria from Unexplored Regions of Western Ghats, India. Microorganisms. 2020;8(2):225.
Crossref - Fasiku SA, Ogunsola OF, Fakunle A, Olanbiwoninu AA. Isolation of Bacteria with Potential of Producing Extracellular Enzymes (Amylase, Cellulase and Protease) from Soil Samples. J Adv Microbiol. 2020;20(3):21-26.
Crossref - Niyonzima FN. Detergent-compatible bacterial cellulases. J Basic Microbiol. 2019;59(2):134-147.
Crossref - Amadi OC, Egong EJ, Nwagu TN, et al. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon. 2020;6(7):e04566.
Crossref - Khusro A, Barathikannan K, Aarti C, Agastian P. Optimization of Thermo-Alkali Stable Amylase Production and Biomass Yield from Bacillus sp. Under Submerged Cultivation. Fermentation. 2017;3(1):7.
Crossref - Tuli DK, Kuila, a. (Eds.). Current Status and Future Scope of Microbial Cellulases. Elsevier. 2021.
- Kumar A, Chandra R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon. 2020;6(2):e03170.
Crossref - Dalagnol LMG, Silveira VCC, da Silva HB, Manfroi V, Rodrigues RC. Improvement of pectinase, xylanase and cellulase activities by ultrasound: Effects on enzymes and substrates, kinetics and thermodynamic parameters. Process Biochem. 2017;61:80-87.
Crossref - Shafique T, Shafique J, Zahid S, Kazi M, Alnemer O, Ahmad A. Screening, selection and development of Bacillus subtilis apr-IBL04 for hyper production of macromolecule alkaline protease. Saudi J Biol Sci. 2021;28(2):1494-1501.
Crossref - Chandra P, Enespa, Singh R, Arora PK. Microbial lipases and their industrial applications: a comprehensive review. Microb Cell Fact. 2020;19(1):169.
Crossref - Manni A, Filali-Maltouf A. Diversity and bioprospecting for industrial hydrolytic enzymes of microbial communities isolated from deserted areas of south-east Morocco. AIMS Microbiol. 2022;8(1):5-25.
Crossref - Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ. Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol. 2020;128(6):1583-1594.
Crossref - Gawas VS, Shivaramu MS, Damare SR, Pujitha D, Meena RM, Shenoy BD. Diversity and extracellular enzyme activities of heterotrophic bacteria from sediments of the Central Indian Ocean Basin. Sci Rep. 2019;9(1):9403.
Crossref - Shahid I, Han J, Hanooq S, Malik KA, Borchers CH, Mehnaz S. Profiling of Metabolites of Bacillus spp. and Their Application in Sustainable Plant Growth Promotion and Biocontrol. Front Sustain Food Syst. 2021;5:605195.
Crossref - Prayogo FA, Budiharjo A, Kusumaningrum HP, Wijanarka W, Suprihadi A, Nurhayati N. Metagenomic applications in exploration and development of novel enzymes from nature: a review. J Genet Eng Biotechnol. 2020;18(1):39.
Crossref - Ahmad T, Gupta G, Sharma A, Kaur B, El-Sheikh MA, Alyemeni MN. Metagenomic analysis exploring taxonomic and functional diversity of bacterial communities of a Himalayan urban fresh water lake. Farooq S, ed. PLoS ONE. 2021;16(3):e0248116.
Crossref - Dong X, Strous M. An Integrated Pipeline for Annotation and Visualization of Metagenomic Contigs. Front Genet. 2019;10:999.
Crossref - Sousa J, Silverio SC, Costa AMA, Rodrigues LR. Metagenomic Approaches as a Tool to Unravel Promising Biocatalysts from Natural Resources: Soil and Water. Catalysts. 2022;12(4):385.
Crossref - Berini F, Casciello C, Marcone GL, Marinelli F. Metagenomics: novel enzymes from non-culturable microbes. FEMS Microbiol Lett. 2017;364(21).
Crossref - Amin K, Tranchimand S, Benvegnu T, Abdel-Razzak Z, Chamieh H. Glycoside Hydrolases and Glycosyltransferases from Hyperthermophilic Archaea: Insights on Their Characteristics and Applications in Biotechnology. Biomolecules. 2021;11(11):1557.
Crossref - Coughlan LM, Cotter PD, Hill C, Alvarez-Ordonez A. Biotechnological applications of functional metagenomics in the food and pharmaceutical industries. Front Microbiol. 2015;6.
Crossref - Verma S, Meghwanshi GK, Kumar R. Current perspectives for microbial lipases from extremophiles and metagenomics. Biochimie. 2021;182:23-36.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.