ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is one of the important opportunistic pathogens causing life threatening nosocomial infections. Both clinical and environmental isolates can be considered potential pathogens due to the conservation of some of the virulence genes. Present study aimed at evaluating the prevalence and diversity of some of the virulence genes among P. aeruginosa isolates from clinical and environmental samples. In this study, 70 clinical (sputum and swab) and 31 environmental isolates were checked for the presence of 8 different virulence genes by PCR amplification. Genetic diversity was studied by performing Enterobacterial repetitive intergenic consensus (ERIC) PCR followed by cluster analysis using GelCompar II software. There was a significant difference in the number of clinical and environmental isolates possessing virulence genes (t = 8.2, c2 = 28.257 at p £ 0.05). 10 clinical and 1 environmental isolate was found to possess all four genes of the type III secretion system (exoS, exoU, exoT and exoY). Cluster analysis revealed 3 major groups with some of the environmental isolates clustering with the clinical ones. To the best of our knowledge, this is the first study to report the presence of all four genes involved in type III secretion system in P. aeruginosa. Presence of several virulence traits in the environmental isolates suggests the possibility of these non clinical ones becoming a clinical pathogenic one. The similarity of three environmental isolates with the clinical ones shows the likelihood of infection caused by them in a hospital setting.
Virulence genes, ERIC PCR, Cluster analysis
P. aeruginosa is considered to be one of the most important causes of nosocomial infections worldwide. Infections with P. aeruginosa remain a major health concern among the immunocompromised and critically ill patients. The success of P. aeruginosa as an effective opportunistic pathogen is partly due to its ability to resist number of antimicrobial agents and also its tolerance to a number of physical conditions. The severity and outcome of infection caused by this pathogen is dependent on the secretion and combination of virulence factors produced 1. Once inside a susceptible individual, it causes host cell death by disrupting host cell signal transduction cascades with a combination of type III secretion effector proteins 2. In P. aeruginosa some of the virulence genes are conserved and this makes both clinical and environmental isolates as potential pathogens 3. Possibility of the environmental isolate becoming a clinical one cannot be ruled out. The differentiation of closely related strains is possible with genomic fingerprinting by PCR amplification using ERIC sequence. ERIC PCR is a successful method to characterize a large number of bacteria4. In India, there are scanty reports on the genotypic characterization of virulence factors from environmental and clinical isolates of P. aeruginosa. With this background, our study aimed at examining the prevalence of virulence genes and its genetic diversity in clinical and environmental isolates of P. aeruginosa.
Isolation of P. aeruginosa
Samples from environmental sources (soil and water) were collected from in and around Mangaluru, India during 2016. P. aeruginosa isolates from clinical exudates (35 sputum and 35 wound swab) collected in 2015 from a tertiary care hospital in Mangaluru and stored as glycerol stocks were used in this study. Environmental samples were selectively enriched using Pseudomonas aspargine broth and selectively isolated in cetrimide agar. Identification of environmental strains was carried out by various biochemical tests (catalase, oxidase, citrate utilization and gelatin liquefaction) as well as by molecular method using species specific primer for the outer lipoprotein (oprL) 5.
Virulence Gene Amplification
DNA from environmental and clinical isolates was subjected to PCR using primers specific for virulence genes. Primers for exotoxinT (5’-3’exoT1: CTCTATACCAACGGCGAGTA and exoT2: TAGAGGATCTCCTGCTCATC) and exotoxinU (5’-3’exoU1: TGATCATCAGCAGTTACTCG and exoU2: TTAGCCATCTCAACGGTAGT were designed using primer3 software. Primers for exotoxin S, exotoxin Y, pyocyanin, alkaline protease, elastase B and pyoverdine were as previously described 5.
ERIC PCR
ERIC sequence from a previous study 6 was used to detect genetic diversity in relation to the virulence genes tested. 101 isolates were subjected for ERIC PCR together with PAO1. Data analysis was performed using GelCompar II software. Band based similarity coefficient was selected as Dice and parameters such as optimization, tolerance were 0.5 % and remaining was zero. UPGMA method was used for the analysis.
Statistical Analysis
The data was subjected to student’s t-test and chi- square test to test for the significance. Data was considered significant at p ≤ 0.05.
Isolation and identification of environmental strains of P. aeruginosa
In total sixty three environmental samples were collected (47 soil and 16 water) of which 51 samples were presumed to be of P. aeruginosa (42 soil and 9 water) based on the initial selective isolation procedure. All 51 isolates were positive for catalase, oxidase, citrate utilization and gelatin liquefaction tests. Upon molecular identification, only 31 isolates (25 soil and 6 water) were confirmed to be of P. aeruginosa. The percent prevalence of confirmed environmental isolates was higher in soil (59%) than in water (21%).
Prevalence of Virulence genes
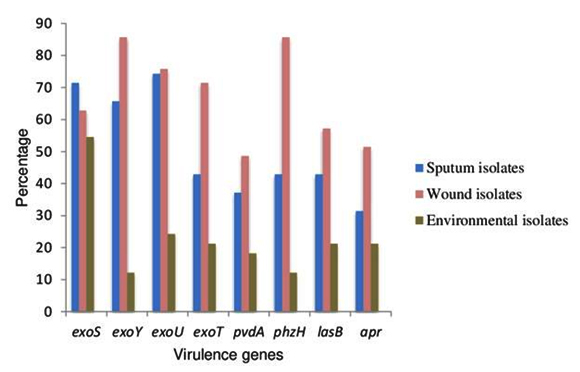
Of the 35 clinical isolates from sputum, 27 isolates were positive for exoY followed by exoS in 25 isolates. Only 11 isolates were found to be harbouring exoT. Of the 35 clinical isolates from wound swab, gene for apr and phz were present in 30 isolates followed by exoY and lasB in 25 isolates. exoU was detected only in 34 isolates out of 70 clinical strains. Eight isolates from sputum and wound swab were found to posses all 8 virulence genes tested. There was a significant difference in the presence of virulence genes in the isolates obtained from different clinical sites [t = 2.07, p ≤ 0.05]. In isolates from water, apr, exoY, lasB and exoT were found to be absent. More number of virulence genes was present in soil isolates than in water isolates. It was revealed that, in majority of the clinical isolates, the percent prevalence of virulence genes was more than 50. But in majority of the environmental isolates, the percent prevalence of virulence genes was less than 25 except for exoS. The percent prevalence of virulence genes is depicted in Fig.1.
Fig. 1. Percent prevalence of virulence genes in clinical and en-vironmental isolates of P. aeruginosa
Student’s t-test revealed significant difference in the number of isolates expressing virulence genes between clinical and environmental ones [t = 8.2, p ≤ 0.05]. There was a significant difference in the presence of virulence genes in the tested isolates [c2 (7, N=103) = 28.257, p ≤ 0.05]. Presence of combination of virulence genes was analysed and data is provided in supplementary material (Fig1). In 10 clinical and 1 environmental isolate there was presence of exoS, exoU, exoT and exoY, the genes involved in type III secretion system.
Genotypic characterization of isolates based on the virulence genes
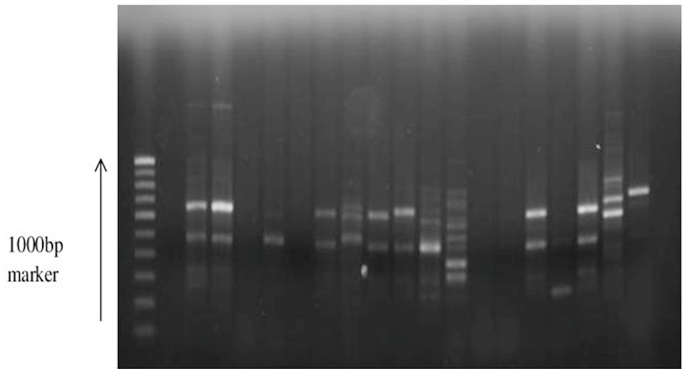
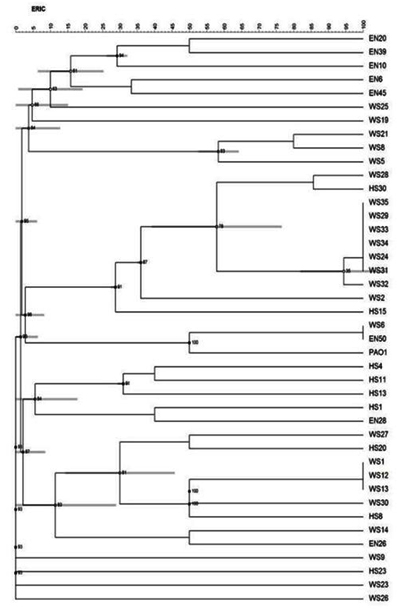
Of the 101 isolates subjected for ERIC PCR, 41 isolates showed positive result (7 environmental and 34 clinical isolates and PAO1) and the remaining were non typable with the sequence used. An analysis of the ERIC banding pattern showed bands varying from 300- 700 bp (Fig. 2). There was no association between the presence of certain virulence genes and any specific pattern. A cluster analysis of the ERIC pattern showed the environmental and clinical isolates clustering into 3 major groups. Most of the environmental isolates clustered together and were distantly placed from the clinical isolates. None of the environmental isolates showed 100% similarity in banding pattern, reflecting the diversity of isolates present in the environment. Three environmental isolates EN50, EN26 and EN28 clustered with clinical isolates (Fig. 3). The clinical isolates were found to be very diverse in their banding pattern with 34 clinical isolates showing 19 different banding patterns. The standard culture PAO1 clustered among the clinical isolates. 9 isolates from wound swab had a similar pattern of alignment at 600 bp (WS 24, 29, 31, 32, 34, 33, 35, 28, and 2); four isolates had a similar pattern of alignment at 500 bp (WS1, WS12, WS13, and HS8). All the remaining isolates had their own unique banding pattern. Maximum diversity was observed in sputum samples.
Selective enrichment on asparagine broth followed by isolation on cetrimide agar which is the method recommended for isolation of Pseudomonas from water and food samples (USFDA BAM, BIS 13856) was used for isolation. Low prevalence of P. aeruginosa in fresh water from Indian coastal regions (~4.1%) has been reported 7. Similar result was reported from Japan 8. In contrast, our study showed a higher prevalence of P. aeruginosa in fresh water samples (21%) from in and around Mangaluru situated in the west coast of India. Higher prevalence in our study might be due to the modified isolation procedure where in enrichment of the sample was carried out with aspergine broth before selective isolation on cetrimide agar.
The percent prevalence of virulence genes in all clinical isolates were more thus making them more virulent causing life threatening infections. Soil isolates with the presence of all the virulence genes tested could possibly express these genes and become pathogenic if they adopt themselves to the hospital environment. An inverse correlation was noted between the presence of exo S and exo U genes. These finding is in accordance with a study which also confirms the inverse correlation between these two genes 9. Mutual exclusivity of exo U and exo S was observed in a previous study 10. The results from our study varied significantly and no mutuality was found to occur between these two genes. The percent occurrence of exo Y gene is not in compliance with a previous study which shows the presence of exo Y in more than 90% of clinical isolates 9. Variation in the production of virulence factors by clinical isolates may be explained by the fact that these isolates are from different clinical sites and collected during different stage of infection 11. Difference in the distribution of virulence genes in clinical and environmental isolates indicates the adaptation mechanisms of this pathogen to different environments 12.
P. aeruginosa isolates harbour at least some of the genes encoding type III secretion apparatus (exoT, exoS, exoU and exoY) and three of the four type III effector proteins are variable. Of the four type III effector genes, vast majority of isolates contain either exoS or exoU gene 13. All four genes of type III effector apparatus has not been described so far in P. aeruginosa isolates. This happens to the first study to reveal the presence of four virulence genes of type III effector genes in both clinical and environmental isolates of P. aeruginosa. Infection with an isolate with type III effector genes has been shown to correlate with severe disease with increased mortality rate 14. Since environmental isolate is also found to carry 4 type III secretion genes, the isolate can be considered more efficient in causing the disease if at all it gains entry into hospital environment.
The use of ERIC PCR has been reported to be a useful technique in confirming the high heterogenicity of isolates 15. Isolates of P. aeruginosa showed a considerable genetic variability, regardless of their habitat. A similarity in the banding pattern of many clinical isolates may be due to similar sampling sites arising from the same source, in this case the hospital. The clustering of three environmental isolates with the clinical ones shows possible relatedness between these isolates.
Presence of several virulence traits in the environmental isolates suggests the possibility of these non clinical ones becoming a clinical pathogenic one. Even though, the percent prevalence of virulence genes in environmental isolates were less compared to the clinical isolates, chances of transmission of such isolates from the environment to the hospital setting cannot be ruled out. Knowledge gained on the prevalence of virulence genes in environmental isolates of P. aeruginosa can improve the existing approaches that target the prevention of transmission from these environmental sources. Further work is being carried out to establish the expression of these genes under suitable environmental condition.
ACKNOWLEDGMENTS
We would like to thank Nitte University for the facilities provided and to Mrs. Swathi S Rao, who was working as a Junior Research Fellow. Funding information: This work was supported by Department of Biotechnology grant (BT/ PR8593/ PFN/ 20/ 763/ 2013).Ethical statement: Ethical clearance was obtained from institutional ethical committee with reference number INST.EC/EC/011/2015-16 on 19.02.2015. Samples collected were anonymized.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med, 2004; 10(12):599-606.
- Lee VT, Smith RS, Tumler B, Lory S. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infection and Immunity, 2005; 73(3): 1695-1705.
- Shaikh S, Fatima J, Shakil S, Rizvi SM. Prevalence of multidrug resistant and extended spectrum beta-lactamase producing Pseudomonas aeruginosa in a tertiary care hospital. Saudi J Biol Sci, 2015; 22(1):62-64.
- Louws FJ, Fulbright DW, Stephens CT, De Brujin FJ. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol, 1994; 60(7):2286-95.
- De-Vos D, Lim A, Pirnay JP, Struelens M. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol, 1997; 35(6) :1295-99.
- Stehling GE, Leite SD, Silveira DW. Molecular typing and biological characteristic of Pseudomonas aeruginosa from cystic fibrosis patients in Brazil. J Infect Dis, 2010; 14(5):462- 467.
- Mohandass C, LokaBharathi P. Representation, dispersion, and variation of bacterial indicators in the coastal waters of Nagore (east coast of India). Water Environ Res , 2003; 75(1):66-72.
- Nonaka L, Inubushi A, Shinomiya H. Differences of genetic diversity and antibiotics susceptibility of Pseudomonas aeruginosaisolated from hospital, river and coastal seawater. Environ Microbiol Rep , 2010; 2(3):465-72.
- Lister PD, Wolter DJ, Hanson ND. Antibacterial resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanism. Clin Microbiol Rev, 2009; 22(4):582-610.
- Kulasekara BR, Kulasekara HD, Wolfgang MC, Stevens L. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J Bacteriol, 2006; 188(11):4037-50.
- Heidary Z, Bandani E, Eftekhary M, Jafari AA. Virulence genes profile of multidrug resistant Pseudomonas aeruginosa from Iranian children with UTIs. Acta Medica Iranica, 2016; 54 (3):201-10.
- Lomholt JA, Poulsen K, Kilian M. Epidemic population structure of Pseudomonas aeruginosa: Evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect Immun, 2001; 69(10):6284-95.
- Feltman H, Schulert G, Khan S, Jain M. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology, 2001; 147(10):2659-69.
- Azimi S, Kafil HS, Baghi HB, Shokrian S. Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect Control, 2016;11.DOI: 10.3205/dgkh000264.
- Olvera A, Calsamiglia M, Aragon V. Genotypic diversity of Haemophilus parasuis field strains. Appl Environ Microbiol, 2006; 72(6):3984-92.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.