Enterococci are gram-positive facultative anaerobes, these are commensals in the intestinal system of numerous animals, including humans. They affect hospitalized patients and cause nosocomial infections, respiratory infections, endocarditis, wound infections, UTIs, and other enterococcal infections. This discovery can be explained by hemolysin, gelatinase, aggregation substances, hyaluronidase, capsular polysaccharides, and cell wall carbohydrate. Various enterococcal spp. include Enterococcus avium Vancomycin resistance was acquired by enterococci throughout the antibacterial spectrum, primary antibiotic used regimen based on dual β lactams and aminoglycosides.
Dual β-lactam’s Aminoglycosides Resistance, Enterococci, Urinary Tract Infections, Bacteremia, Endocarditis Antibiotic
Enterococci are Gram-positive catalase-negative, non-spore-forming, facultative anaerobic lactic acid bacteria and normal inhabitants of the gut flora of humans, many different mammals, birds, fish, reptiles, amphibians and insects, as well as nematodes.1
Enterococci, which are gram-positive facultative anaerobes, are present commensals in the digestive tracts of numerous animals, including humans.2 They are also present in the soil, water, dairy foods, and even plant, primarily present on the mucosal surfaces of people and animals. Because these illnesses have the potential to spread human pathogens to hospitalized patients, they are referred to as nosocomial infections.1 UTIs and wounds in bacteremia bacteremias are all examples of enterococci infections. They frequently come with endocarditis, pelvic infections, and intra-abdominal infections. Otitis, sinusitis, septic arthritis, endophthalmitis, and pathogens infections of the nose, throat, and brain can also happen.3
Many Enterococcus species [105–108 colony-forming units] coexist in the wild as nosocomial pathogens and have grown in recent decades. The natural bacterial ecology in both human and animal intestines is made up of gram-positive, facultative anaerobic cocci called Enterococcus species. A high death rate and protracted hospitalization are two effects of enterococcal bacteraemia.1-3 The two most common Enterococcal spp. that cause bacteremia is Enterococcus faecalis and Enterococcus faecium.4
These two enterococci species most frequently isolated and linked to infections contracted in hospitals are Enterococcus faecalis and Enterococcus faecium. Sequencing has been done on the genomes of E. faecalis and E. faecium.5 There is only one VRE strain, Faecalis strain V583, that exhibits the Van B trait A pathogenicity island, several recombinant and composition -displacement, integration plasmids sequences, phage sections, a significant amount of insertion sequences and about 25 percent of the genome are mobility or extracellularly acquired DNA (IS).6 134 potential surface-exposed proteins that may be connected to virulence or colonization were discovered by a comprehensive genome-wide investigation.7
Virulence factors
Numerous enterococci strains that were antibiotic-resistant were first identified in the 1970s, except those that cause endocarditis infections. Up until the discovery of numerous multidrug resistances in the 1970s, enterococci were thought to be innocuous bacteria, except those that may cause endocarditis. There has been an increase in enterococcal nosocomial pathogens among hospitalized patients during the past 20 years.8 These specifically happened in intensive care settings, which are conducive hosts for bacterial colonization outside of the body. UTIs occasionally lead to GIT, which can result in abdominal injury. Numerous nosocomial illnesses resulting in the production of Intent carditis bacteremia have discovered enterococcal surface protein is one of many bacterial pathogens (Esp.) gelatinase, hemolysin, aggregation substance (AS).
Hemolysin
It is a cytolytic protein that can cause lysis in both people and horses and red blood cells from rabbits. Enterococci strains that cause hemolysis have been demonstrated to be hazardous to animals and humans. Increased infection severity hemolysis occurrence can be recognized by exposing oneself to newly manufactured enterococci. Horse blood should be infused into the Beef Heart Infusion Agar along with Ove. Plates were kept at 37°C in a carbon dioxide chamber for 24 hours of incubation; distinct hemolysis zones around colonies on horse blood agar it is regarded as positive.7 Cytolysin or hemolysis-dependent regulation of expression a novel quorum sensor-based two-component regulatory system.9
Gelatinase
A protease produced by enterococci that may hydrolyze peptides such as collagen, gelatine, casein, and hemoglobin. E. faecalis strains that produce gelatinase have been connected to the virulence of endocarditis in animal models. By inoculating freshly made gelatine plates with enterococci on peptone yeast extract agar, incubation at 37°C Overnight, and then chilling to room temperature for two hours, it is possible to measure the generation of gelatinase in a laboratory setting positive for the synthesis of gelatinase.10
Aggregation Substances
It is an E. faecalis pheromone-induced surface protein that promotes the development of mating aggregation during bacterial conjugation.9 Aggregation Substance (AS) effectively enhances the transfer of plasmids between enterococcal donors and recipients. Through a variety of methods, aggregation material may be involved in the development of enterococcal infection.10 In experiments using PCR amplification it was proven that there are no asa-type genes (encoding for the aggregation ingredient). Additionally, asa1- and asa373-specific gene probes.11
Hyaluronidase
When K-hyaluronate was provided in a suitable agar medium, it was shown that SF68 did not produce hyaluronidase because it was not broken down. Further evidence of the gene’s absence was provided by PCR and a bioinformatics search inside the SF68 genomic sequence.12
MSCRAMM ace
The structural and functional similarities between staphylococcal Cna adhesion and the collagen-binding adhesin generated by enterococci, or “ace,” are striking.12 (MSCRAMM, or microbial surface component recognizing adhesive matrix molecule). Its occurrence in pathogenic and commensal E. faecalis isolates.13
Cell wall and capsular polysaccharide
Clinical isolates of E. faecalis have been reported to express an operon that codes for the production of a specific type of capsular polysaccharide.14 A 14-second experiment utilizing a mouse infection model that showed the protective efficiency of antibodies produced against this refined carbohydrate component suggested the notion that these antibodies may help prevent enterococcal infections.15 it was discovered that pure cellular glycogen components contained residues of glycol phosphatase, fructose, or lactate.
Emergence and Dissemination Trends of Vancomycin-Resistant Enterococci (VRE) Strains
Vancomycin-resistant enterococci (VRE) have spread with unanticipated rapidity and today are steadily increasing worldwide. However, European countries and the United States (US) have experienced differences in VRE emergence and epidemiology.16 In the 1990s, the rapid emergence of VRE observed in the US was preceded by the emergence of ampicillin-resistant E. faecium in the early 1980s. At the same time, in Europe, the first VRE clinical isolates were only detected in 1986.17 Although the proliferation of VRE strains currently posing a major threat to human infection management, vancomycin is still a commonly used antibiotic to treat infections brought on by multi-resistant enterococci.18
Control of vancomycin Resistant Enterococci
Controlling VRE dissemination in pediatric patients requires prompt detection of VRE by microbiology laboratories, education of staff and families about VRE, use of infection control measures to prevent person-to-person VRE transmission, and prudent vancomycin use.19
The Public Health Impact of VRE
Historically, E. faecalis was responsible for the majority of all enterococcal infections (80-90%); however, in recent years, the proportion of E. faecium infections has surpassed that of E. faecalis.20
Other Species of Enterococci
E. avium
Gram-positive, catalase-negative streptococcus known as E. avium is frequently isolated from birds. Previously, group Q streptococcus 16 was the name for E. avium even though these bacteria were known to cause bacteriemia and thus had the potential to cause other chronic infections, there aren’t many reports about its involvement in human illnesses.21 Only two cases of bacterial meningoencephalitis caused by E. avium have been reported, both of which had a successful result but did not involve persistent otitis media.22 This condition appears to entail a different pathogenetic mechanism than brain abscesses. Ovoid cells elongated along the chain, usually in pairs or short chains. Non-motile: On blood or nutrient agar, surface colonies are circular, smooth, and complete. Nonpigmented: On blood agar, the majority of strains produce an alpha-reaction.9
E. durians
The gastrointestinal tract’s natural flora includes E. durians. Few case reports have been documented since Sherman and Wing discovered this rare Enterococcus species in human pathology in 1935. This is because there are few described pathogenic factors and low virulence of E. durians, like other species of the enterococcus genus, has positive results for the group D antigen in the Lancefield classification system.16 It is stationary and does not require mannitol as a source of energy, in contrast to E. avium, E. raffinoses, and other enterococcus species.
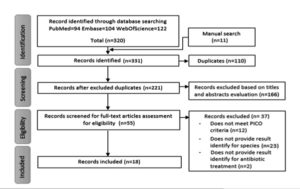
Amino acids and B vitamins are required for growth in synthetic media, which is nutritionally demanding. It thrives on 0.1% methylene blue milk. Growth was not observed in media containing 0.04% tellurite and 0.01% tetrazolium. H2S is not formed.17 Figure denotes the study selection Flow chart reporting screening of the database for eligibility criteria.18
E. raffinoses
Enterococcus raffinose, the most mysterious of the three species discussed in this research, was just recently separated. It is negative for tellurite and arginine but positive for mannitol, sorbose, arabinose, raffinose, and pyruvate.23 Biological Profile 2022, 11, 598, and this biochemical profile Due to 13 of the 17 species’ immobility, identification is exceedingly challenging in labs lacking an automatic detection tool The potential for E. raffinoses to possess the vanA gene, which confers resistance to glycopeptides, has been questioned. Recent research has shown that vanA outbreaks are indeed occurring in a variety of locations, making this OE extremely hazardous to the medical community and even to the general public health.24
Enterococcus gallinarum
Coccoid cells, mostly in pairs or short chains. Nonmotile. Colonies on blood agar or nutrient agar are circular, smooth, and entire. Beta-hemolytic on horse blood agar.10 Nonpigmented: Most strains do not survive 30 minutes of heating at 60°C, but do survive 15 minutes of heating at 60°C.8 At room temperature, it grows slowly on thallous acetate-tetrazolium agar, producing deep pink colonies.10 L-arginine produces ammonia. Gelatin has not been liquefied. There is no H2S produced.25
Review
Holzapfel et al. in studied He conducted assays to determine the presence of virulence factors such as hemolysin, gelatinase, hyaluronidase, endocarditis antigen, and aggregation material Because no adhesion was seen to any of the surfaces investigated, these results supported the notion that SF68 has a very limited ability to adhere to intestinal epithelial cells. The effectiveness and safety of pharmaceutical probiotics were evaluated using Enterococcus faecium SF68 as a model.26
In 2020, Laura Herrera-Hidalgo and others, among the study types utilized in the 18 publications that were selected were randomized clinical trials (n = 1), non-randomized clinical trials (n = 1), prospective cohort studies (n = 2), retrospective cohort study (n = 9), series study (n = 5). Only 10 of them looked at several potential treatments. Outpatient (n = 4), inpatient (n = 9), and mixed (n = 5) clinical settings were used for the investigation, nine studies included continuation therapy as a therapeutic indication. Except for three research that solely addressed left-sided IE, the bulk of investigations included both left- and right-sided endocarditis. There were significant differences in the follow-up time after the antimicrobial therapy ended, ranging from no follow-up20,27
Similarly, in 2020 according to Sayed Hossein Mousavi et al., who studied the PCR results, 59 (53.2%) and 25 (22.5%) of the 111 clinical isolates were E. faecalis and E. faecium, respectively. A total of 60.3%, 56.7%, and 51.35 percent of the isolates were (HLG) & (HLS), respectively. Thirteen (48.14%), 18 (72%), and 36 (61.01%) of the HLGR isolates were non-fecal non-faecium species, E. faecalis respectively. Among the HLSR isolates, E. faecalis, E. faecium, and non-fecal non-faecium species were represented by 33 (55.93%), 16 (64%), and 14 (51.85%), respectively. Every HLGR isolate had the aac (6′) Ie-aph (2′′) Ia gene. 17.1% of Enterococcus species have high levels of ampicillin resistance overall. Ampicillin resistance rates for E. faecalis, E. faecium, and non-fecal non-faecium spp. 11 (40.74%), 7 (28%), and 1 (1.69%), respectively, antibiotics called aminoglycosides.28
Alexander et al. in worked on Enterococcus raffinoses, Enterococcus durians, and Isolates of Enterococcus avium that were collected from the Romanian tertiary-care hospital. A (retrospective) study found that he performed antibiotic sensitivity by Vitek 2 compact and obtained 658 isolates of enterococcus involved in humans. Of these, 319 isolates, or 48%, were excluded because they were not identified using an automated vitek system.29 From the remaining isolates, E. faecalis made up 126 (37.16%), E. faecium 155 (45.72%), and other enterococci made up 58 (17.10%). The strains were selectively isolated from hospital wastewater and then identified using matrix-assisted laser desorption and ionization time-of-flight mass spectrometry. Antibiotic susceptibility testing was performed using the disc diffusion method.30
Hence our study highlights the significance of virulence factors that are responsible for the pathogenicity of enterococci species, as this led to increasing multidrug resistance accounting for more hospital-acquired infections. Since some drugs are intrinsically resistant to Enterococci and mobile genetic elements are used to spread resistance to other bacteria. So, detection is based on conventional gram staining and culture methods, in addition to molecular methods such as MALDI-TOF, NAAT, and PCR. In comparison to older procedures, modern ones are believed to be more dependable and sensitive; therefore, this article examines how to diagnose and treat enterococcal infections.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Escobedo-Hinojosa W, Pardo-Lopez L. Analysis of bacterial metagenomes from the Southwestern Gulf of Mexico for pathogens detection. Pathog. Dis. 2017;75(5):1-9.
Crossref - Franz CM, Muscholl-Silberhorn AB, Yousif NM, Vancanneyt M, Swings J, Holzapfel WH. Incidence of virulence factors and antibiotic resistance among Enterococci isolated from food. Appl Environ Microbiol. 2001;67(9):4385-4389.
Crossref - Padmasini E, Padmaraj R, Ramesh SS. High-level aminoglycoside resistance and distribution of aminoglycoside resistant genes among clinical isolates of Enterococcus species in Chennai, India. Scientific World Journal. 2014;329157.
Crossref - Bhardwaj S, Dhawale KB, Patil M, Divase S. Enterococcus faecium and Enterococcus faecalis, the nosocomial pathogens with special reference to multi-drug resistance and phenotypic characterization. International Journal of Pharmaceutical Science and Practice. 2013;2(1):1-10.
- Donskey CJ, Chowdhury TK, Hecker MT, et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engle J Med. 2000;343(26):1925-1932.
Crossref - Sherman JM, Wing HU. Streptococcus durians n. sp. Journal of Dairy Science. 1937;20(3):165-167.
Crossref - Collins MD, Jones D, Farrow JAE, Kilpper-Balz R, Scleifer KH. ‘Enterococcus avium nom. rev., comb. Nov.; E. Casseliflavus Nom. rev., comb. Nov.; E. Durans Nom. rev., comb. Nov.; E. Gallinarum Comb. Nov.; and E. Malodoratus sp. nov..’, International Journal of Systematic Bacteriology, 1984;34(2):220–223.
Crossref - Farrow JA, Jones D, Phillips BA, Collins MD. Taxonomic studies on some group D streptococci. J Gen Microbiol. 1983;129(5):1423-1432.
Crossref - Miller WR, Murray BE, Rice LB, Arias CA. Resistance in vancomycin-resistant enterococci. Infect Dis Clin North Am. 2020;34(4):751-71.
Crossref - Bridge PD, Sneath PH. Streptococcus gallinarum sp. nov. and Streptococcus oralis sp. nov. Int J Syst Evol Microbiol. 1982;32(4):410-415.
Crossref - Barnes EM, Mead GC, Impey GS, Adams BW. The effect of dietary bacitracin on the incidence of Streptococcus faecalis subspecies liquefaciens and related streptococci in the intestines of young chicks. British Poultry Science. 1978;19(6):713-723.
Crossref - Deibel RH, Lake DE, Niven CF Jr. Physiology Of The Enterococci as Related to their Taxonomy. J Bacteriol. 1963;86(6):1275-1282.
Crossref - Ruoff KL, de la Maza L, Murtagh MJ, Spargo JD, Ferraro MJ. Species identities of enterococci isolated from clinical specimens. J Clin Microbiol. 1990;28(3):435- 437.
Crossref - Patel R, Keating MR, Cockerill FR, Steckelberg JM. Bacteremia due to Enterococcus avium. Clin Infect Dis. 1993;17(6):1006-1011.
Crossref - Holzapfel W, Arini A, Aeschbacher M, Coppolecchia R, Pot B. Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef Microbes. 2018;9(3):375-388.
Crossref - Hayakawa K, Marchaim D, Vidaillac C, et al. Growing Prevalence of Vancomycin-Resistant Enterococcus faecalis in the Region with the Highest Prevalence of Vancomycin-Resistant Staphylococcus aureus. Infection Control & Hospital Epidemiology. 2011;32(9):922-924.
Crossref - Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008;52(3):297-308.
Crossref - Marino A, Munafò A, Zagami A, et al. Ampicillin Plus Ceftriaxone Regimen against Enterococcus faecalis Endocarditis: A Literature Review. J Clin Med. 2021;10(19):4594. Published 2021 Oct 6.
Crossref - Upadhyaya PMG, Ravikumar KL, Umapathy BL. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol. 2009;27(4):301-305.
Crossref - Rich RL, Kreikemeyer B, Owens RT, et al. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274(38):26939-26945.
Crossref - Huebner J, Quaas A, Krueger WA, Goldmann DA, Pier GB. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect Immun. 2000;68(8):4631-4636.
Crossref - Toc DA, Pandrea SL, Botan A, et al. Enterococcus raffinose, Enterococcus durans a, and Enterococcus avium Isolated from a Tertiary Care Hospital in Romania-Retrospective Study and Brief Review. Biology. 2022;11(4):598.
Crossref - Jolivet S, Fines-Guyon M, Nebbad B, et al. First nosocomial outbreak of vanA-type vancomycin-resistant Enterococcus raffinose in France. 2016;94(4):346-350.
Crossref - Herrera-Hidalgo L, de Alarcon A, Lopez-Cortes LE, et al. Enterococcus faecalis Endocarditis and Outpatient Treatment: A Systematic Review of Current Alternatives. Antibiotics. 2020;9(10):657.
Crossref - Cattoir V. The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr Opin Microbiol. 2022;65:73-80.
Crossref - Ramos S, Silva V, Dapkevicius MLE, Igrejas G, Poeta P. Enterococci, from Harmless Bacteria to a Pathogen. Microorganisms. 2020;8(8):1118.
Crossref - Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. Comparative genomics of vancomycin-resistant Enterococcus spp. revealed common resistome determinants from hospital wastewater to aquatic environments. Sci Total Environ. 2020;719:137275.
Crossref - Cherak Z, Bendjama E, Moussi A, et al. First detection of vanA positive Enterococcus faecium clonal complex 17 in hospital wastewater in Algeria: an epidemiological report. New Microbes New Infect. 2022;47:100977.
Crossref - Shay DK, Goldmann DA, Jarvis WR. Reducing the spread of antimicrobial-resistant microorganisms. Control of vancomycin-resistant enterococci. Pediatr Clin North Am. 1995;42(3):703-716.
Crossref - Markwart R, Willrich N, Haller S, et al. The rise in vancomycin-resistant Enterococcus faecium in Germany: data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob Resist Infect Control. 2019;8:147.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.