ISSN: 0973-7510
E-ISSN: 2581-690X
Pigeon peas are an excellent source of carbohydrates, proteins and other nutrients. Many traditional fermented foods are prepared from cereals and combinations of cereals and pulses that usually contain Lactic acid bacteria (LAB), Bacillus, Enterococcus and yeast. Lactobacillus can be used as a starter culture for such fermentation using pulses, as very few reports are available on fermented pulse-based products. Hence, pulse dal flour was used as a source for isolation of Lactobacillus to maintain their functionality, growth characteristics and activity during food processing. In this study, we investigated the potential of lactobacilli from fermented pigeon pea to utilize carbohydrates, the ability to degrade non-digestible oligosaccharides and the production of the α-galactosidase enzyme. Lactobacillus isolated from six different pulse dal flour grew well during fermentation with carbohydrates in mMRS medium. Among Lactobacillus species, only Lactobacillus brevis displayed the highest α-galactosidase activity (1.24 U/ml), where raffinose was added as the sole carbohydrate source in the medium. The isolate was further tested in pigeon pea fermentation, where it showed maximum activity (1.86 U/ml) and complete hydrolysis of non-digestible oligosaccharides was observed. Overall, usage of Lactobacilli could be an excellent opportunity to design and develop a novel pulse-based fermented product contributing to beneficial bioactive compounds and improving the properties of food.
Cajanus cajan, Lactobacillus, Fermentation, Carbohydrate utilization, Non-digestible oligosaccharides, α-galactosidase (α-gal) activity
Pigeon pea (Cajanus cajan) also known as tuvar, red gram, and arhar belonging to the leguminosae family, are consumed in various forms like biscuits, pasta, noodles, sausages as well as fermented product like Tempe throughout the world. They are rich in carbohydrates (65%) and protein (20-25%) and comprise of basic nutritive constituents including dietary fibers, minerals, vitamins, amino acids, fatty acids and phytochemicals.1,2 Cajanus cajan contributes to various health benefits in treating diabetes, reproductive system infections, stabilizing menstrual issues, skin irritation, additionally biological properties like antioxidant, antibacterial, antitumor, anti-inflammatory are associated widely.3,4 The main drawback limiting the consumption of pigeon pea is the presence of high level of α-galactosides, mainly raffinose that comprises of sugars with galactose units, linked as α-1, 6-galactosyl residue. Humans are deficient in producing α-galactosidase (α-gal), as a result it is not digested in intestine which leads to abdominal pain, flatulence, diarrhea, nausea, etc.5 Therefore, to overcome this problem attempts have been made over the years to eliminate this galactoside from pigeon pea to enhance nutritional value of a food product. Thus, the use of bacteria producing α-galactosidase offers a promising solution for the degradation of this oligosaccharide during pigeon pea fermentation by Lactobacillus. Currently, various Lactobacilli such as L. fermentum, L. plantarum, L. reuteri, L. helveticus, L. acidophilus and L. brevis are capable of hydrolyzing galactosides into palatable form.6,7 For elimination of these galactosides, fermentation is one such method that helps in improving physicochemical and functional properties of fermented food.
Lactobacilli have been used as the starter culture for their contribution to longer shelf life, flavor and aroma. A number of studies revealed several biotherapeutic values of lactobacilli in fermented food including degrading toxic moiety and fortifying health-promoting bioactive components.8 Various fermented foods using cereals are popularly prepared in India and other parts of the world but very few pulse-based fermented food products such as kinema, dhokla currently exist in the market. Using Lactobacillus as a carrier in pulses will serve food of high calorie, improved functionality and sensory qualities. Several groups have suggested the presence of lactobacilli in various bean based products employing fermentation in the form of sourdough or beverage.9,10 However to the best of our knowledge, no study has addressed culturing of lactobacilli in pulse dal flour. In the present study, we determined the microbial ability to degrade sucrose and non-digestible oligosaccharide (NDO), raffinose via fermentation of pigeon pea using Lactobacillus isolates.
Chemicals and strain
The chemicals used for the analysis were purchased from HiMedia Laboratories Pvt. Ltd. (Mumbai, Maharashtra, India), Sisco Research Laboratories Pvt. Ltd. (Mumbai, Maharashtra, India) and Merck Life Science Private Limited (Mumbai, Maharashtra, India). Lactobacillus plantarum 1407T a type strain, isolated from fermented cabbage was procured from Microbial Type Culture Collection and Gene Bank (MTCC) (Chandigarh, India). This culture was used as reference strain for comparison with obtained isolates and was preserved on MRS slants at 4°C.
Preparation and fermentation of pulse dal flour
Dal such as tuvar (Cajanus cajan), masoor (Lens culinaris), vaal (Vicia faba), udad (Vigna mungo), chana (Cicer arietinum), and mung (Vigna radiate) were purchased from a local market in Vadodara, Gujarat, India and milled to fine size flour using mortar and pestle. The flours were stored at room temperature until further use in an airtight container. In a sterile container, flour was mixed with sterile deionized water in 1:2 (w/v) ratio and volume of batter was marked before fermentation. In a similar way, fermentations were carried out for the rest of the flours and batters were mixed thoroughly. Volume of the batter before fermentation was measured and batters were kept at 37°C for 24 h to obtain lactobacilli. Fermentations were propagated by naturally present microorganisms in the flour and samples were collected at 12 h and 24 h for analysis.
Microbiological analysis, characterization and identification of Lactobacillus spp.
Ten grams of each sample was suspended in 90 ml of sterile 0.8% (w/v) saline, homogenized with vortex for 30 seconds and serially diluted. Lactobacilli were isolated at 37°C in MRS (de Man, Rogosa, Sharpe) after 2-3 days. Based on the cell morphology, colonies were picked and transferred to fresh MRS plates and were examined for catalase production and only those colonies were selected for Gram’s staining which were catalase negative. Identification of Lactobacillus at species level was done according to Bergey’s Manual of Determinative Bacteriology, and tested for carbohydrate fermentation.11 Phenotypic characterization was done on the basis of growth at different temperatures (10, 15 and 45°C), pH values (4.5 and 9.0) and NaCl concentrations (0.5, 1, 2, 4 and 6.5% (w/v)) in MRS broths.12 The purified strains of lactobacilli were maintained on MRS slants at 4°C until further use.
For genotyping characterization of isolates, samples were sent to Labreq Bioscientific, Ahmedabad, Gujarat, India. Genomic DNA isolation followed by 16s rRNA sequencing was done using BigDyeTM Terminator Cycle Sequencing Kit on a 3730xl DNA Analyzer (Applied Biosystems, CA, USA). A GeneBank database search of the partial 16S rRNA sequences was conducted to determine the closest relatives of the bacterial sequence using BLAST algorithm.13
Utilization of carbohydrates
Single and mixed combinations of carbohydrates like sucrose, soluble starch + sucrose, raffinose, and raffinose + soluble starch were selected as carbon sources. The experiment was conducted in 35 ml using modified MRS (mMRS) medium (g/l) composed of 1 g yeast extract, 2 g ammonium citrate, 5 g sodium acetate, 2 g dipotassium hydrogen phosphate, 0.1 g magnesium sulphate, 0.05 g manganese sulphate, 1 ml tween 80 per litre and sugars: 1% sucrose, 0.3% starch + 0.5% sucrose, 1% raffinose, 1% raffinose + 0.3% starch were suspended separately in different Erlenmeyer flask and sterilized by autoclaving at 121°C for 15 min. 1 ml of Lactobacillus culture (12 log cfu/ml) was inoculated and grown in respective sugar medium at 37°C overnight. Samples were withdrawn at every 4 h of fermentation, for monitoring microbial counts, pH drop, decrease in sugars, etc. For comparison, reference strain was inoculated in the similar way as described above. Control was an uninoculated sample for blank.
Chemical analysis
Based on the cell–count, growth characteristics were monitored at different time intervals. In an Erlenmeyer flask each fresh culture were inoculated at a final concentration of 108 cfu/ml in 35 ml of mMRS broth medium followed by incubation at 37°C at every 4 h interval up to 12 h. Optical density was recorded at 600nm in an UV-visible 1800 spectrophotometer (Shimadzu, Japan). 2 ml of sample was removed from each isolate at every 4 h. Cell supernatant was collected by centrifuging at 10,000 x g for 10 min and pH was determined.
Lactic acid in the samples were determined using spectrophotometric method.14 Starch content in the sample was analyzed by iodine- starch assay and the blue color developed was measured at 590 nm.15 The concentration of sucrose and raffinose during fermentation were determined by spectrophotometric method at 432.5 nm.16 The production of glucose during fermentation was estimated using dinitrosalicylic acid method and absorbance was measured at 540 nm.17
Extracellular enzyme activity
A loopful of culture was inoculated in sterilized mMRS medium, followed by addition of filter sterilized pNPG (p- nitrophenyl-α-D-galactopyranoside) and incubated at 37°C for 48 h. The formation of color indicated the production of α-gal enzyme. In the assay condition, 1U of enzyme was defined as the amount of enzyme that would liberate 1.0 µmol of p– nitrophenol from the substrate pNPG per ml per min. Cell growth was evaluated and protein was determined using bovine serum albumin as standard.18
Characterization of α-gal activity
To test the effect of pH, enzyme was kept at different pH ranging from 2 to 11. Similarly, the optimal temperature of α-gal activity was determined ranging from 0 to 60°C.
The growth medium composition was evaluated for the production of α-gal activity using different carbon sources like raffinose (0.5%, 1%), starch (0.3%, 0.5%, 1%) and sucrose (0.5%, 1%). The preparation of the enzyme assay was performed similar to the method described above.
Fermentation using pulse beans
To evaluate the fermentation behavior, 10 g of whole pigeon pea beans were soaked in water (10:40 g:ml) overnight (12 h), rinsed twice with fresh water, followed by autoclaving at 121°C for 15 min, dehulled and kept for fermentation at 37°C for 48 h. The culture was activated till it reached the stationary phase in MRS medium to a final OD600nm= 5. Cells were centrifuged at highest speed for maximum 10 min. Fermented pigeon pea were sampled at set intervals to check for microbial population, decrease in pH, hydrolysis in raffinose and sucrose using thin layer chromatography,16 α-gal activity, and lactic acid.
Statistical analysis
All results are expressed as mean ±standard deviation and performed in triplicates. Statistical significance was analyzed using one-way analysis of variance (ANOVA). The data with p < 0.05 was considered significant.
Sequences accession number
The sequences were deposited under GeneBank NCBI (http://www.ncbi.nlm.nih.gov/) with following accession numbers: M1: MK530231; U1: MK530234; VIP1: MK530235; TIP1: MK530232.
Isolation of Lactobacillus from fermented pulse dal flour
The first strategy was aimed to carry out dal flour fermentation for the isolation of Lactobacillus. The fermentation set up for all the flour batters is presented in Table 1. After 24 h of propagation, the microbial population recorded on MRS medium ranged from 3.00-5.00 log cfu/ml at 37°C with pH 4.5 at the end of fermentation. No bacterial growth was detected during the initial hours of fermentation. As the fermentation progressed, it was observed that the pH of the batter was decreasing, and lactobacilli population was increasing. Negligible growth of other bacteria was observed at the end of fermentation probably due to low pH of the batter. A large increase in the population of yeast and a rise in the volume of batter was observed after 24 h of fermentation. It has been reported that the role of yeast can be directly correlated to the rise in the volume of dal batter and CO2 production.19 The microbial flora present in spontaneous fermentation of sourdough includes many diverse species of microorganism such as Lactococcus, Weissella, Leuconostoc, Pediococcus, Streptococcus and Enterococcus, while lactobacilli are the most frequently observed bacteria during fermentation.20 There are certain other popular fermented food products like ogi, kenkey, busaa, and koko, a cereal and maize based food items showed mainly the presence of Lactobacillus genera involved in the process.21 In general, Lactobacillus are considered the predominant organism in fermented food.22
Table (1):
Analysis of pulse dal flour during fermentation.
Sample (designated) |
Vol. of batter before fermentation (ml) at 0 h |
Vol. of batter after fermentation (ml) after 24 h |
pH of batter after 24 h |
LAB count (log cfu/ml) after 24 h |
No. of isolates |
|---|---|---|---|---|---|
Masoor dal (M1) |
50 |
90 |
4.5 |
5 |
1 |
Udad dal (U1) |
50 |
200 |
4.5 |
5.07 |
1 |
Tuvar dal (T1, TIP1) |
50 |
80 |
4.5 |
4.30 |
2 |
Val dal (V1, VIP1) |
50 |
85 |
4.5 |
5.04 |
2 |
Mung dal (Mu1) |
50 |
90 |
4.5 |
3.14 |
1 |
Chana dal (C1) |
50 |
80 |
4.5 |
5 |
1 |
“cfu” colony-forming unit
Screening of the isolates
A total of eight bacterial isolates were obtained from dal flour based on their distinct cell morphology on MRS agar. The size of the colonies varied from 0.1 to 0.5 mm in diameter and appeared singly or in clusters. All of these isolates were Gram positive and tested negative for catalase, citrate, urease, Voges-Proskauer and H2S production (Table 2). Four of the isolates metabolized glucose fermentatively in Hugh and Leifson medium and produced gas from glucose, therefore, deemed to be heterofermentative organism. Out of the eight isolates obtained, four isolates were lactobacilli and the remaining belonged to Weissella group. These isolates showed resistance to temperature, saline stress and acid tolerance. It was observed that presumed Lactobacillus could grow at 20 and 45°C. This high temperature can reduce the chances of contamination by other organisms.23 These organisms also managed to survive at 0.5% NaCl concentration but not above. Earlier report indicated low pH in Kunu, an acid-fermented beverage by lactobacilli species resulting in production of organic acid by fermentation of sugars.24 Similarly isolated lactobacilli have also shown tolerance to low pH (~4.0) probably due to its adaptation ability to acid at the time of their presence in fermented dal flour. Therefore NaCl could help in selecting Lactobacillus spp. as a starter culture in salt containing food, ultimately leading to good quality product and longer shelf life.
Table (2):
Characteristics and identification profile of the isolates.
| Isolates/Characteristics | M1 | U1 | VIP1 | TIP1 |
|---|---|---|---|---|
| Cell morphology | Medium, round, entire, capitate, opaque and creamy | Medium, round, entire, convex, opaque and creamy | Medium, round, entire, capitate, opaque and creamy | Small, round, entire, convex, opaque and white |
| Gram’s staining | +, long rod | +, long rod | +, long rod | +, short rod |
| Catalase | – | – | – | – |
| Carbohydrate metabolism | ||||
| Glucose | + | + | + | + |
| Xylose | + | + | + | + |
| Galactose | + | + | + | + |
| Fructose | + | + | + | – |
| Mannitol | + | + | + | – |
| Lactose | + | + | + | – |
| Maltose | + | + | + | + |
| Starch | + | + | + | + |
| Sucrose | + | + | + | + |
| Raffinose | + | + | + | + |
| Co2 production from glucose | + | + | + | + |
| Fermentative type | Hetero | Hetero | Hetero | Hetero |
| Growth at: | ||||
| Temperature range 20 ̊C – 45 ̊C |
+ | + | + | + |
| Lowest pH tolerance | 4.0 | 4.0 | 4.0 | 4.0 |
| NaCl tolerance range 0.5% | + | + | + | + |
| H2S production | – | – | – | – |
| Voges-Proskauer | – | – | – | – |
| Methyl red test | + | + | + | + |
| Urease test | – | – | – | – |
| Citrate reduction | – | – | – | – |
| Identification of isolates through National Centre for Biotechnology Information (NCBI) | ||||
| Identification probabilities (%) | 99 | 99 | 99 | 100 |
| Identity | Lactobacillus plantarum | Lactobacillus pentosus | Lactobacillus plantarum | Lactobacillus brevis |
+, positive; –, negative; hetero- heterofermentative
The source of the isolates are mentioned in table 1.
Genetic identification and phylogenetic tree of Lactobacillus isolates
Based on all the morphological, physiological and biochemical characteristics of all these four isolates listed in Table 2, identification was carried out at species level by 16S rRNA sequencing method. The search in the database was performed in GenBank NCBI using the BLAST program, exhibited similarity up to 99% to 100%. Sequencing data showed that the presumed Lactobacillus clearly showed similarity to Lactobacillus plantarum, Lactobacillus pentosus, Lactobacillus plantarum and Lactobacillus brevis isolated from masoor dal (M1), udad dal (U1), val dal (VIP1), and tuvar dal (TIP1) respectively.
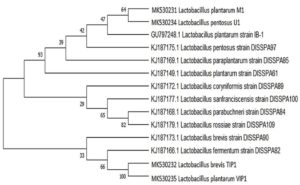
A phylogenetic tree was generated from sequenced 16S rRNA region of four bacterial isolates identified from fermented dal flour batter compared with reference strain isolated from idli batter and pulses flour sourdough.25,26 The evolutionary analysis was constructed using Neighbor Joining method by BLAST program with 1,000 bootstrap values in MEGA X as well as the relationship between Lactobacillus isolates and different reference strains (Fig. 1). The phylogenetic tree built on 16S rRNA sequence indicated that L. plantarum M1 and L. pentosus U1 isolate shares similarity with L. plantarum IB-1 strain. It also revealed that there is a very close genetic relationship between L. brevis, TIP1 and L. plantarum, VIP1. This clade showed near relation to L. fermentum strain DISSPA82. Whilst, L. plantarum M1 and L. pentosus U1 showed a long-distance relationship with other isolates. The relatedness can be inferred from the bootstrap value among the isolates and strains given at the branch.
Fig. 1. Phylogenetic tree of Lactobacillus isolates. It was inferred using Neighbor-Joining method showing the position of isolates and related strains based on 16S rRNA gene sequence. The number at the starting of each genus indicates an accession number obtained from the NCBI database. The evolutionary distance were computed using the Masximum composite Likehood method. The evolutionary analysis was conducted in MEGA X.
Utilization of carbohydrates by Lactobacillus spp.
Pulses are rich source of carbohydrates, constituting a major portion of dry matter27,28 and contains starch, a storage carbohydrate in good amount along with sucrose, the most abundant sugar present in pulses, followed by raffinose, stachyose and verbascose constituting up to 70-75% of total carbohydrates.29,30 Previous reports have shown that most of the Lactobacillus spp. possess starch metabolism pathway while most of them can metabolize sucrose which is a highly preferred substrate.31 Moreover, few strains of lactobacilli can also degrade raffinose through sucrose-hydrolyzing enzyme.31 Therefore, a strategy was developed to determine the efficiency of Lactobacillus isolates to be able to metabolize the carbohydrates by fermentation. For this, synthetic medium containing added carbohydrate nutrients such as starch, sucrose and raffinose, were selected. The concentration of these carbohydrates was based on their average quantity present in pulses. Residual growth was observed in mMRS medium when tested in presence of yeast extract while omitting other organic nitrogen sources. Here attention was given in developing low-cost medium with high efficiency production yield.
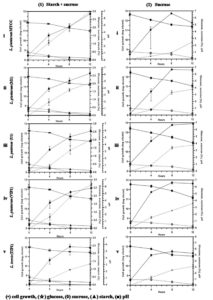
All the five isolates grew well in the tested carbon source. Fig. 2A-1 (i-v) shows the classic bacterial growth pattern of isolates, plotted against time in mMRS media supplemented with 0.3% starch + 0.5% sucrose. The log phase was observed from 4 to 8 h followed by growth leveling off for some isolates while the die off phase started for some after 8 h in the fermentation experiment. The total bacteria in fermentation by lactobacilli consistently increased from 8.00 to 11.50 log cfu/ml and the highest was 12.90 log cfu/ml in 12 h fermentation. An easy availability to excess nutrients in a medium, tends microbes to grow faster and multiply in higher numbers. However, bacteria number in fermentation decreased to 12.69 and 11.77 log cfu/ml at 8 h and 12 h, respectively. The decrease seen in bacterial count that inhibited the microbial growth is perhaps due to over accumulation of metabolites such as lactic acid, acetic acid, ethanol and carbon dioxide; increase in the number of lactobacilli is associated with the capacity to utilize carbon source.32 Several authors reported that fermentation decreases the pH resulting in acidity and increase in lactic acid accumulation by microbial activity.32-34 Similarly, in all the sugars which we tested, increase in cell growth led to decrease in pH from 6.5 to 4.0 indicating an association between pH and growth. Thus it can be sensed that lactic acid production lowers the pH of the media as the growth of lactobacilli is enhanced. All isolated lactobacilli can be tagged as amylolytic due to their ability of utilizing starch as literature also confirms their species of utilizing and producing lactic acid by direct conversion.32,35-38 The highest consumption of starch in the medium was found in U1 and L. plantarum MTCC culture (62%) followed by M1, VIP1, and TIP1 (50%) after 12 h of fermentation (Fig. 2A-1 (i-v)). The result showed that this Lactobacillus spp. metabolized starch during fermentation, indicating the degradation of starch as fermentation progressed. Fermentation activates starch hydrolyzing enzymes and degrades starch to low molecular weight forms, and sugars like monosaccharides, disaccharides and dextrins.39-41 The rise in the glucose level during fermentation was stated due to starch degradation by amylase, while subsequent reduction in sugar content could be because of utilization of a carbon source by fermenting microorganisms.40
Fig. 2A. (Kinetics of different carbohydrate utilization using starch + sucrose (1), and sucrose (2), during fermentation with Lactobacillus isolates and reference strain in mMRS medium).
In this study, sucrose utilization (Fig. 2A-2 (i-v)) was investigated with an initial concentration of 1%. Fermentation was performed at 37°C, which enhanced the microbial population, along with decreasing pH. The use of initial sucrose concentration of 1% resulted in complete sugar consumption within 12 h at final pH between 4-4.8 for all the isolates. The utilization of sucrose is due to the presence of invertase enzyme in the organism that breaks down sucrose to glucose and fructose through catabolism. Granito et al. observed diminution of sucrose and increase of glucose level during natural fermentation of beans.42 Several reports have shown that these Lactobacillus spp. indicated good growth on media containing sucrose43 as well as utilizing sucrose from various other sources such as cane sugar44 and sugarcane juice.45 Thus utilization of sucrose from a low cost source undergoes a promising process for acidic fermentation.
Fig. 2B. (Kinetics of different carbohydrate utilization using raffinose (1), and raffinose + starch (2), during fermentation with Lactobacillus isolates and reference strain in mMRS medium).
This Lactobacillus spp. was further investigated for their potential growth in mMRS medium containing raffinose (1%) and fermented as previously mentioned (Fig. 2B-1 (i-v)). All the isolates were successively able to grow on mMRS-raffinose medium, and rapid utilization of raffinose was observed. All lactobacilli showed almost similar growth capacity on mMRS broth, reaching maximum to 11.57 – 12.10 log cfu/ml after 12 h of incubation depending on the lactobacilli. The acidification capacity during growth was excellent for most isolates, lowering the pH value to less than 4.5 (~4.0-4.2) after 12 h of incubation. Findings of this study are in line with a previous report which showed that raffinose was substantially consumed by L. plantarum.46-48 A probiotic L. pentosus strain was isolated from naturally fermented alorena green table olives exhibited capacity to ferment raffinose but not stachyose.49 A report of L. brevis and its strain isolated from caper berry fermentations showed no utilization of raffinose in synthetic medium.50 M1, VIP1, U1 and TIP1 including reference strain were successfully able to reduce raffinose (p<0.05) by ~80%.
The growth pattern based on the colony count, pH, and utilization of carbon source were obtained for all the isolates growing in media supplemented with 1% raffinose + 0.3% starch (Fig. 2B- 2 (i-v)). The initial inoculum volume was the same for all the isolates. The number of bacterial count increased rapidly for the all lactobacilli including the reference strain and achieved a similar density at stationary phase above 12.00 log cfu/ml; thereafter the cell count fell off more rapidly. Fig. 2B-2 (i-v) indicates that utilization of raffinose and starch by lactobacilli was simultaneous and achieved the same viable cell count for all the isolates when both sugars were present in the medium.
Overall, Lactobacillus isolated from dal flour showed excellent metabolism of starch, sucrose and raffinose as the sole carbon source in the medium. At the end of fermentation, lactic acid production was on par with fermentation using other carbohydrate sources. There was no significant difference (p<0.05) in various carbohydrates added to mMRS medium. However, fermentation time showed a significant effect on the population, carbohydrate utilization and sugar formation from 0 to 12 h.
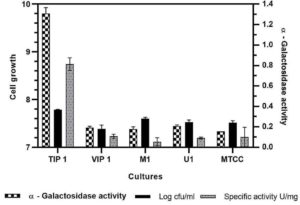
Screening of α-galactosidase activity and cell growth in Lactobacillus isolates
All Lactobacillus isolates and a reference strain were evaluated for the production of α-gal activity and cell growth (Fig. 3)). The microbial growth obtained was in range of 7.38-7.78 log cfu/ml (VIP1 showing 7.38 log cfu/ml while TIP1showed 7.78 log cfu/ml). TIP1 culture showed the highest α-gal activity of 1.30 ± 0.05 U/ml compared to other isolates and the specific activity of protein was 0.81 ± 0.06 U/mg. In this study, the extracellular fraction had considerably higher enzyme activity than the intracellular fraction (data not shown). These results indicated that the microbial enzymes are either intracellular or extracellular.51-53 Previous literature have reported that most of the α-gal are intracellular enzymes with few strains showing both extracellular and cell bound form.7 However, in this study the results have shown higher extracellular enzyme activity. Although, it is an advantage that this enzyme is extracellular, as it provides high yield, stability and broad-pH range in activity compared to intracellular enzymes.54-56 Furthermore, this isolate was selected for more detailed research on NDO and α-gal activity as it has not been investigated yet from fermented pulse dal flour source isolate, L. brevis (TIP1).
Fig. 3. Evaluation of a-galactosidase activity, cell growth and specific activity for different Lactobacillus isolates in mMRS medium.
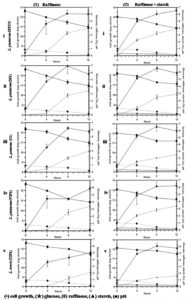
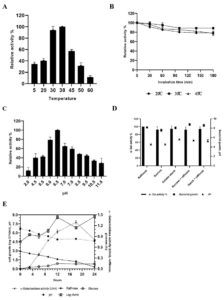
Effect of temperature and pH on α-galactosidase activity by TIP1
The effect of temperature on α-gal activity when incubated with substrate at different temperatures is shown in Fig. 4A, B. The results indicated increase in α-gal activity with hike in temperature, showing maximum activity at 38°C. However, rapid decrease in enzyme activity was observed as the temperature increased, probably due to thermal inactivation6. These results match with the previous reports of α-gal activity by L. acidophilus and L. helveticus at 37°C, while L. plantarum and L. fermentum have been reported to show activity at 50°C. 57-61 Initially, the enzyme showed temperature stability between 0-45°C, and still retains its activity (Fig. 4B). The enzyme maintained about 80% of the stability at 45°C (Fig. 4B), afterwards loss in activity was seen at 50°C.
Fig. 4. Effect of temperature (A), temperature stability (B), pH (C); effect of different carbon source in growth medium (D) on the production of a-galactosidase activity and kinectics of a-galactosidase, bacterial growth and hydrolysis of raffinose (E) during fermentation with L. brevis in mMRS medium.
The effect of pH on the production of α-gal enzyme by TIP1 is depicted in Fig. 4C. α-Gal activity was observed in pH range between 2-11; however, the maximum enzyme secretion was observed at pH 6.5. Negligible amount of enzyme activity was detected in the alkaline range. Similar results have been obtained by G.Tzortzis et al. from L. reuteri where maximum α-gal production was reported in acidic condition.62
Effect of different carbon source on α-galactosidase production ability of TIP1
The effect of different carbon sources was examined on the production of α-gal activity and bacterial growth (Fig. 4D). To induce bacterial galactosidase, all sugars present in pulses were tested for maximum enzyme production. Raffinose expressed the strongest induction, followed by starch, starch + raffinose, sucrose and sucrose + raffinose. No considerable difference in the relative induction levels for other sugars were observed, especially when used in combination. The maximum enzyme production was induced in the presence of raffinose, which confirms the observation in L. plantarum.63 The studied Lactobacillus showed the highest enzyme production when the concentration was 5.0 g/l, while no influence in enzyme activity was observed when the concentration was increased by 30.0 g/l (data not shown). Also, no noticeable difference was seen in the cell growth and pH reduction in growth medium with different types of carbon source.
Time course of α-galactosidase production in TIP1
The kinetics of TIP1 showing α-gal activity, cell densities and degradation product of raffinose due to hydrolysis were determined at different time in mMRS medium (Fig. 4E). It grew very rapidly during the initial hours, reaching the maximum growth of 8.06 ± 0.28 log cfu/ml after 12 h of fermentation and then declined slowly. The mMRS medium pH dropped from 6.70 ± 0.0 to 4.60 ± 0.1 after 24 h of culture inoculation. Meanwhile, α-gal activity appeared in the medium broth within the initial hours, which indicates that the cell is utilizing available carbon source, reaching to a maximum activity of 1.29 U/ml after 18 h of fermentation. These results were in agreement with Garro et al., where similar phenomenon was observed with α-gal activity, following the same growth pattern of the cells and degradation of raffinose.64 On the other hand, it was observed that the consumption of raffinose increases α-gal activity, decrease in activity was noted over the course of time. At the end, raffinose decreased to a non-detectable level after 24 h of fermentation.
Hydrolysis of NDO and α-galactosidase activity in pigeon pea bean fermentation supplemented with TIP1
Hydrolysis of NDO and α-galactosidase activity in pigeon pea bean fermentation supplemented with TIP1
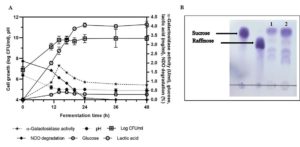
The α-gal activity of TIP1 was assessed during fermentation including its growth and potential in utilizing oligosaccharide for development of biofunctional fermented pigeon pea bean product. NDO are considered the most important oligosaccharides present in pigeon pea, and are believed to cause disturbance in humans after eating. These oligosaccharides are further metabolized by microflora, yielding considerable amounts of carbon dioxide, methane and hydrogen gas in the large intestine.63 This Lactobacillus species showed excellent growth and highest α-gal activity during the start of stationary phase in bean fermentation (Fig. 5A). TIP1 culture showed the highest log count between 0-14 h of incubation under the conditions used in this study. The cell started to multiply immediately after inoculating it in medium for fermentation and soon increased from 6.89 log cfu/ml to 9.78 log cfu/ml after 14 h of incubation. The growth of lactobacilli in initial hours of fermentation might be due to the active log culture used at the time of inoculation and better availability of carbohydrates such as starch, raffinose, sucrose, etc.49 Moreover, higher initial content of carbohydrates facilitate lactobacilli to survive for maximum hours in the medium. After 14 h of incubation, there was almost no growth observed; once the organism entered stationary phase it remained constant till the end of 48 h. The pH of the medium decreased from 6.3 to 5.0 after 14 h, indicating the growth performance of the culture during bean fermentation. Due to enhanced acidification capacity by TIP1, reduction in pH was observed during fermentation.
Fig. 5. Change in pH reduction, bacterial growth, residual sugars, lactic acid and a-galactosidase activity during fermentation with pigeon pea bean by L. brevis (A). Chromatogram (TLC) separation pattern of non-digestible oligosaccharides in untreated (control) and L. brevis treated pigeon pea bean. Lane 1: 18-h-treated pigeon pea, Lane 2: untreated pigeon pea bean (B). Experiment was done in triplicates.
Utilization of sucrose and raffinose in pigeon pea fermentation varied and appeared to be based on the α-gal activity of culture. TIP1 reduced NDO concentration significantly (P < 0.05) by 2.10 to 0.10 mg/100ml after 24 h of incubation at 37°C. α-Gal is the principal enzyme necessary to break the galactoside bond present in sucrose and raffinose; these oligosaccharides are the primary substrate for the enzyme producing lactobacilli. Interestingly, α-gal activity was higher in case of TIP1 studied in pigeon pea fermentation compared to activity in mMRS medium supplemented with pNPG substrate. The highest value of α-gal activity was 1.86 U/ml at the initial exponential phase and sharply decreased by the end of the fermentation. The total NDO content in the unfermented pigeon pea was found to be 24.0 g/l. In the fermentation medium a complete disappearance of oligosaccharide occurred by 36 h; whereas increase in enzyme activity was detected as the time progressed. Meanwhile another peak corresponding to glucose appeared and progressively remained throughout fermentation. α-Gal activity is a species and strain dependent process that has been documented in many lactobacilli. Further confirmation in reduction of oligosaccharides (sucrose, raffinose) was done using thin layer chromatography (TLC) under chromatographic conditions mentioned by Tanaka et al.16 TLC analysis of fermented beans using α-gal secreted by L. brevis, showed hydrolysis of sucrose and raffinose after 18 h of incubation (Fig. 5B). TLC chromatogram did indicate the reduction of NDO in organism treated bean sample as against their level in untreated sample (control). At the same time, two more bands appeared on TLC plate which could possibly be verbascose and stachyose due to their higher carbohydrate content in pigeon pea after sucrose and raffinose.29 As documented in previous studies with fermented cereals as well as legumes, fermentation using GRAS organisms could effectively lower down galacto-oligosaccharides due to presence of higher α-gal enzyme.65 Lactobacilli are known to impart several benefits to the product due to their special characteristics59; these benefits were often studied using enzymatic treatment containing high NDO.66 Several reports in literature confirms use of α-gal enzyme isolated from Aspergillus niger65 and Bacillus megaterium5 for removal of raffinose family from cowpea flours and soymilk. Here, the advantage of using microorganism over the bacterial α-gal enzyme is its cost effectiveness. Thus, it would be promising by using native culture with more than one benefit in removing flatulence causing oligosaccharides and imparting probiotic features to the product. Four lactobacilli species were selected for further study in pigeon pea fermentation and at the end only one was found to have potential for maximum production of enzyme α-galactosidase.
Lactobacillus is one of the most common GRAS organisms recognized in fermented food and thus it is important to address its effect in reducing NDO using different strains during preparation of pulse-based product. Hence, using this Lactobacillus could be a good opportunity for using a single desirable culture in maintaining consistent quality and developing a novel fermented pulse-based product with microbial safety, improved flavors as well as appropriate in absorption capacities of consumers. However, the present study did not convey the anti-nutritional properties in pigeon pea fermentation by TIP1. Therefore, further investigation is required to study the reduction of the antinutritives such as tannins, saponins during fermentation with pigeon pea.
ACKNOWLEDGMENTS
I am thankful for the assistance provided by the Department of Microbiology and Biotechnology Centre, Vadodara for their contribution in achieving the work. I would also like to thank Manasi Patil and Naisarg Gamit for their help in reviewing the article.
FUNDING
This study was supported by financial aid from University Grants Commission (UGC)-BSR (Grant No. F.25-1/2014-15(BSR)/7-128/2007(BSR)).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Rathinam M, Mishra P, Vasudevan M, et al. Comparative transcriptome analysis of pigeonpea, Cajanus cajan (L.) and one of its wild relatives Cajanus platycarpus (Benth.) Maesen. PLoS One. 2019;14(7):e0218731.
Crossref - Talari A, Shakappa D. Role of pigeon pea (Cajanus cajan L.) in human nutrition and health: A review. Asian J Dairy Food Res. 2018;37(3):212-220.
Crossref - Jiao J, Gai Q-Y, Wang X, et al. Effective Production of Phenolic Compounds with Health Benefits in Pigeon Pea [Cajanus cajan (L.) Millsp.] Hairy Root Cultures. J Agric Food Chem. 2020;68(31):8350-8361.
Crossref - Pal D, Mishra P, Sachan N, Ghosh AK. Biological activities and medicinal properties of Cajanus cajan (L) Millsp. J Adv Pharm Technol Res. 2011;2(4):207-214.
Crossref - Patil AGG, Kumar SK P, Mulimani VH, Veeranagouda Y, Lee K. α-Galactosidase from Bacillus megaterium VHM1 and Its Application in Removal of Flatulence-Causing Factors from Soymilk. J Microbiol Biotechnol. 2010;20(11):1546-1554.
Crossref - Carevic M, Banjanac K, Corovic M, et al. Selection of lactic acid bacteria strain for simultaneous production of α-and β-galactosidases. Zastita materijala. 2016;57(2):265-273.
Crossref - De Cort S, Kumara HS, Verachtert H. Localization and characterization of α-glucosidase activity in Lactobacillus brevis. Appl Environ Microbiol. 1994;60(9):3074-3078.
Crossref - Tamang JP, Shin D-H, Jung S-J, Chae S-W. Functional properties of microorganisms in fermented foods. Front Microbiol. 2016;7:578.
Crossref - Hueda MC. Functional Food: Improve Health through Adequate Food. TechOpen, London. 2017.
Crossref - Saez GD, Hebert EM, Saavedra L, Zarate G. Molecular identification and technological characterization of lactic acid bacteria isolated from fermented kidney beans flours (Phaseolus vulgaris L. and P. coccineus) in northwestern Argentina. Food Res Int. 2017;102:605-615.
Crossref - Bergey DH, Breed RS, Murray EGD, Hitchens AP. Manual of determinative bacteriology, 5th Ed. Williams & Wilkins Co., Baltimore. 1939.
- Sengun IY, Nielsen DS, Karapinar M, Jakobsen M. Identification of lactic acid bacteria isolated from Tarhana, a traditional Turkish fermented food. Int J Food Microbiol. 2009;135(2):105-111.
Crossref - Huttner EK, Dal Bello F, Arendt EK. Identification of lactic acid bacteria isolated from oat sourdoughs and investigation into their potential for the improvement of oat bread quality. Eur Food Res Technol. 2010;230:849-857.
Crossref - Borshchevskaya L, Gordeeva T, Kalinina A, Sineokii S. Spectrophotometric determination of lactic acid. J Anal Chem. 2016;71:755-758.
Crossref - McGrance SJ, Cornell HJ, Rix CJ. A simple and rapid colorimetric method for the determination of amylose in starch products. Starke. 1998;50(4):158-163.
Crossref - Tanaka M, Thananunkul D, Lee T-C, Chichester CO. A simplified method for the quantitative determination of sucrose, raffinose and stachyose in legume seeds. J Food Sci. 1975;40(5):1087-1088.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426-428.
Crossref - Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1-2):248-254.
Crossref - Sridevi J, Halami PM, Vijayendra S. Selection of starter cultures for idli batter fermentation and their effect on quality of idlis. J Food Sci Technol. 2010;47(5):557-563.
Crossref - Corsetti A, Settanni L. Lactobacilli in sourdough fermentation. Food Res Int. 2007;40(5):539-558.
Crossref - Petrova P, Petrov K. Lactic acid fermentation of cereals and pseudocereals: Ancient nutritional biotechnologies with modern applications. Nutrients. 2020;12(4):1118.
Crossref - Rezac S, Kok CR, Heermann M, Hutkins R. Fermented foods as a dietary source of live organisms. Front Microbiol. 2018;9:1785.
Crossref - Menconi A, Kallapura G, Latorre JD, et al. Identification and characterization of lactic acid bacteria in a commercial probiotic culture. Biosci Microbiota Food Health. 2014;33(1):25-30.
Crossref - Ayo-Omogie H, Okorie E. In vitro probiotic potential of autochthonous lactic acid bacteria and microbiology of Kunu made from mixed grains. Microbiol Res J Int. 2016;14(4):1-10.
Crossref - Iyer BK, Singhal RS, Ananthanarayan L. Characterization and in vitro probiotic evaluation of lactic acid bacteria isolated from idli batter. J Food Sci Technol. 2013;50(6):1114-1121.
Crossref - Rizzello CG, Calasso M, Campanella D, De Angelis M, Gobbetti M. Use of sourdough fermentation and mixture of wheat, chickpea, lentil and bean flours for enhancing the nutritional, texture and sensory characteristics of white bread. Int J Food Microbiol. 2014;180:78-87.
Crossref - Robinson G, Balk J, Domoney C. Improving pulse crops as a source of protein, starch and micronutrients. Nutr Bull. 2019;44(3):202-215.
Crossref - Singh N. 4-Functional and physicochemical properties of pulse starch, Pulse Foods. 2nd Ed. Academic Press, USA. 2011, 91-119.
Crossref - Venkidasamy B, Selvaraj D, Nile AS, Ramalingam S, Kai G, Nile SH. Indian pulses: A review on nutritional, functional and biochemical properties with future perspectives. Trends Food Sci Technol. 2019;88:228-242.
Crossref - Singh B, Singh JP, Shevkani K, Singh N, Kaur A. Bioactive constituents in pulses and their health benefits. J Food Sci Technol. 2017;54(4):858-870.
Crossref - Ganzle M, Follador R. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol. 2012;3:340.
Crossref - Pranoto Y, Anggrahini S, Efendi Z. Effect of natural and Lactobacillus plantarum fermentation on in-vitro protein and starch digestibilities of sorghum flour. Food Biosci. 2013;2:46-52.
Crossref - Wedad HA, El Tinay AH, Mustafa AI, Babiker EE. Effect of fermentation, malt-pretreatment and cooking on antinutritional factors and protein digestibility of sorghum cultivars. Pak J Nutr. 2008;7(2):335-341.
Crossref - Ibrahim FS, Babiker EE, Yousif NE, El Tinay AH. Effect of fermentation on biochemical and sensory characteristics of sorghum flour supplemented with whey protein. Food Chem. 2005;92(2):285-292.
Crossref - Abriouel H, Perez Montoro B, Casimiro-Soriguer CS, et al. Insight into potential probiotic markers predicted in Lactobacillus pentosus MP-10 genome sequence. Front Microbiol. 2017;8:891.
Crossref - Sanni A, Morlon-Guyot J, Guyot J. New efficient amylase-producing strains of Lactobacillus plantarum and L. fermentum isolated from different Nigerian traditional fermented foods. Int J Food Microbiol. 2002;72(1-2):53-62.
Crossref - Akin-Osanaiye B, Azeez B, Olobayotan I. Evaluation of Invertase and Amylase Activities of Latic Acid Bacteria Isolated from Pupuru (An Indigenous African Fermented Cassava Staple Food). Asian J Res Biochem. 2019;5(3):1-8.
Crossref - Rahayu E, Hidayah N, Adiandri R. Profile of Modified Sorghum Flour Fermented by Lactobacillus Brevis. Paper presented at: IOP Conf. Ser. Earth Environ. Sci. 2019.
Crossref - Nkhata SG, Ayua E, Kamau EH, Shingiro JB. Fermentation and germination improve nutritional value of cereals and legumes through activation of endogenous enzymes. Food Sci Nutr. 2018;6(8):2446-2458.
Crossref - Abd Elmoneim OE, Bernhardt R, Bonomi F, Iametti S, Pagani MA, Zardi M. Fermentation modifies protein/protein and protein/starch interactions in sorghum dough. Eur Food Res Technol. 2006;222:559-564.
Crossref - Osman MA. Effect of traditional fermentation process on the nutrient and antinutrient contents of pearl millet during preparation of Lohoh. J Saudi Soc Agric Sci. 2011;10(1):1-6.
Crossref - Granito M, Alvarez G. Lactic acid fermentation of black beans (Phaseolus vulgaris): microbiological and chemical characterization. J Sci Food Agric. 2006;86(8):1164-1171.
Crossref - Ounis WB, Champagne C, Makhlouf J, Bazinet L. Utilization of tofu whey pre-treated by electromembrane process as a growth medium for Lactobacillus plantarum LB17. Desalination. 2008;229(1-3):192-203.
Crossref - Gomaa EZ, Rushdy AA. Improvement of Lactobacillus brevis NM101-1 grown on sugarcane molasses for mannitol, lactic and acetic acid production. Ann Microbiol. 2014;64:983-990.
Crossref - Timbuntam W, Sriroth K, Tokiwa Y. Lactic acid production from sugar-cane juice by a newly isolated Lactobacillus sp. Biotechnol Lett. 2006;28(11):811-814.
Crossref - Fritsch C, Vogel RF, Toelstede S. Fermentation performance of lactic acid bacteria in different lupin substrates-influence and degradation ability of antinutritives and secondary plant metabolites. J Appl Microbiol. 2015;119(4):1075-1088.
Crossref - Mital BK, Steinkraus KH. Fermentation of soy milk by lactic acid bacteria. A review. J Food Prot. 1979;42(11):895-899.
Crossref - Donkor ON, Henriksson A, Vasiljevic T, Shah N. α-Galactosidase and proteolytic activities of selected probiotic and dairy cultures in fermented soymilk. Food Chem. 2007;104(1):10-20.
Crossref - Montoro BP, Benomar N, Lavilla Lerma L, Castillo Gutierrez S, Galvez A, Abriouel H. Fermented Alorena table olives as a source of potential probiotic Lactobacillus pentosus strains. Front Microbiol. 2016;7:1583.
Crossref - Pulido RP, Omar NB, Abriouel H, et al. Characterization of lactobacilli isolated from caper berry fermentations. J Appl Microbiol. 2007;102(2):583-590.
Crossref - Li Y, Sun L-L, Sun Y-Y, et al. Extracellular enzyme activity and its implications for organic matter cycling in northern Chinese marginal seas. Front Microbiol. 2019;10:2137.
Crossref - Traving SJ, Thygesen UH, Riemann L, Stedmon CA. A model of extracellular enzymes in free-living microbes: which strategy pays off? Appl Environ Microbiol. 2015;81(21):7385-7393.
Crossref - Reintjes G, Arnosti C, Fuchs B, Amann R. Selfish, sharing and scavenging bacteria in the Atlantic Ocean: a biogeographical study of bacterial substrate utilisation. ISME J. 2019;13(5):1119-1132.
Crossref - Burns RG. Microbial extracellular enzymes and natural and synthetic polymer degradation in soil. Molecular Environmental Soil Science. Springer. 2013:22-47.
Crossref - Chauhan AS, Kumar A, Siddiqi NJ, Sharma B. Extracellular α-Galactosidase from Trichoderma sp.(WF-3): Optimization of Enzyme Production and Biochemical Characterization. Biotechnol Res Int. 2015;2015:860343.
Crossref - Katrolia P, Rajashekhara E, Yan Q, Jiang Z. Biotechnological potential of microbial α-galactosidases. Crit Rev Biotechnol. 2014;34(4):307-317.
Crossref - Fredslund F, Abou Hachem M, Larsen RJ, et al. Crystal structure of α-galactosidase from Lactobacillus acidophilus NCFM: insight into tetramer formation and substrate binding. J Mol Biol. 2011;412(3):466-480.
Crossref - Kandari S, Choi YJ, Lee BH. Purification and characterization of hydrolytic and transgalactosyl α-galactosidase from Lactobacillus helveticus ATCC 10797. Eur Food Res Technol. 2014;239(5):877-884.
Crossref - Roopashri AN, Varadaraj MC. Hydrolysis of flatulence causing oligosaccharides by α-d-galactosidase of a probiotic Lactobacillus plantarum MTCC 5422 in selected legume flours and elaboration of probiotic attributes in soy-based fermented product. Eur Food Res Technol. 2014;239(1):99-115.
Crossref - Garro MS, de Valdez GF, Oliver G, de Giori GS. Purification of α-galactosidase from Lactobacillus fermentum. J Biotechnol. 1996;45(2):103-109.
Crossref - Carrera-Silva EA, Silvestroni A, LeBlanc J, Piard J-C, de Giori GS, Sesma F. A thermostable α-galactosidase from Lactobacillus fermentum CRL722: genetic characterization and main properties. Curr Microbiol. 2006;53(5):374-378.
Crossref - Tzortzis G, Jay A, Baillon M, Gibson G, Rastall R. Synthesis of α-galactooligosaccharides with α-galactosidase from Lactobacillus reuteri of canine origin. Appl Microbiol Biotechnol. 2003;63(3):286-292.
Crossref - Silvestroni A, Connes C, Sesma F, De Giori GS, Piard J-C. Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl Environ Microbiol. 2002;68(11):5464-5471.
Crossref - Garro MS, de Valdez GF, de Giori GS. α-Galactosidase assay in fermented soymilk products. Environ Microbiol. 2004:121-124.
Crossref - Girigowda K, Prashanth S, Mulimani V. Oligosaccharins of black gram (Vigna mungo L.) as affected by processing methods. Plant Foods Hum Nutr. 2005;60(4):173-180.
Crossref - Kapnoor S, Mulimani VH. Production of α-galactosidase by Aspergillus oryzae through solid-state fermentation and its application in soymilk galactooligosaccharide hydrolysis. Braz Arch Biol Technol. 2010;53(1):211-218.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.