ISSN: 0973-7510
E-ISSN: 2581-690X
Currently, microalgae have become a marvelous and resource-friendly alternative source of advantageous bioproducts, such as lipids, carbohydrates, proteins, or other bioactive compounds. Because of the richness of microalgae in these high-value-added metabolites, still, it is an underdeveloped source of sustainable energy and food. There are some hurdles to profitable production, such as culture contamination and costly harvesting techniques. In the current work, a chrysophyte was isolated from dairy wastewater, identified as Poterioochromonas malhamensis based on its morphology and partial 18S rRNA gene sequences. This isolate was used to remediate dairy waste water (DWW) and to obtain neutral lipids (fatty acids) from microalgae. Microalgal growth was optimized by using different concentrations of DWW, supplemented with all the nutritive requirements for better progression and flourishment. Maximum biomass yield 1.478 g L−1 was achieved by optimized cultural conditions (different concentrations of DWW with BBM media). This strain showed high nitrate and phosphate removal efficiency (87.45% and 88.96%), respectively in 15 days. The experimental results highlighted that the lipid content and the chemical oxygen demand (COD) removal were 31.60% and 88.84%, respectively, and the lipid profile of isolated microalga was C16:0, C16:1, C18:0, C18:1, and C18:2 fatty acids. For growth and treatment purposes, 75% DWW with Bold’s Basal Medium (BBM) media showed better results. This is the first report of DWW treatment using the microalga Poterioochromonas malhamensis, as far as we are aware. Its cultivation prevented the spread of pollution of freshwater sources, remedied the DWW, and generated important lipids for industry.
Dairy Wastewater, Chrysophyte, Poterioochromonas malhamensis, Bioremediation, Lipid Production
In the current scenario, an escalating population and rapid growth of industries, overexploit the natural resources of water and produce a huge volume of wastewater so the availability of fresh water has become a major global challenge. Untreated or heavily contaminated wastewater released into the environment has adverse effects on the normal operations of ecosystems, flora, and fauna, which make the world under a serious dilemma. Different types of industries produce different types of waste water, among these, the dairy industry is one of them and produces a huge amount of waste water in the form of strong pollutants (organic and inorganic compounds).1 The dairy industry has played a significant role in the Indian economy and provides great job prospects, further; this industry has been growing day by day. The dairy industry has an advantageous impression at the country’s financial expansions, although, being the largest wastewater or effluent developer. According to FAO 2020, India has become the largest milk producer in the world with 22% of global milk production followed by the USA, and Pakistan. The dairy industries produce huge amounts of wastewater and globally, it has a deep ecological impact. These industries guzzle a lot of water when compared with other agricultural industries. It has been evaluated that the processing of one liter of milk consumes approximately three liters of freshwater.2 Major dairy product i.e., milk has reported a great expansion in manufacturing and utilization. Across the world, the utilization of dairy products would have been expected to enhance up to 13.7% by 2023.3-5 The most complicated issue raised by discharging untreated dairy wastewater into the freshwater resources, the exhaustion of oxygen in water, hence increasing biological oxygen demand. Dairy wastewater has higher amount of grease or fat molecules thus it creates a superficial layer on waterbodies which obstructs the transfer of oxygen and creating difficulty for the survival of flora and fauna present there.3,6 Dairy wastewater is also rich in organic compounds so it helps to intensify the growth of microbes and causes eutrophication in freshwater resources if drained without proper treatment. Dairy effluent is non-toxic in nature when compared with other industrial discharges.7 Various physicochemical techniques, such as screening, sedimentation, chemical precipitation, oxidation, flocculation, coagulation processes, filtration, flotation, chlorination, neutralization, electrochemical and coagulation, absorption, ion exchange, etc., have been applicable in the treatment of wastewaters. But these have several restrictions including expensive, incomplete treatment, production of sludge and xenobionts, etc.8 Large quantities of sludge and the use of harsh chemicals make the physicochemical techniques unsuitable for the eradication of these contaminants from dairy effluent. There is strict governance on clear disposal, which has administered the need for possible other innovational and feasible procedures for dairy waste prevention.9-11
To deal with an emerging concern about dairy waste remediation and environmental protection, alternatively based on the microalgae cultivation suggests a prospective future.12 Due to having the surprising capability to multiply on organic waste and producing beneficial substances, thus microalgae, are a self-sustainable alternative that can simultaneously provide a solution to not only remediation of waste water but also to be useful in bioenergy production. That’s why achieved great recognition from researchers to culture them extensively for the generation of biomolecules and wastewater treatment. Employing mother nature’s “green gold” to decontaminate waste water while also manufacturing sustainable food and other valuable byproducts is assumed to be an easily accomplished task. This green technology for bioremediation of pollutants encompassing advanced technology related to controlling of environmental pollution is in vogue.13
Microalgae offer several advantages such as rapid growth rate, efficient carbon dioxide sequestration, removal of nutrients, oxygen production, and treat dairy waste simultaneously producing biomass which contains various valuable organic bioactive compounds like lipids, proteins, carbohydrates, pigments, antioxidants, and vitamins, etc. these organic compounds, have wide applications in the field of human health, nutraceuticals, cosmetics, pharmaceuticals, and biofuels also, 5,11,14 and most importantly, they do not have any adverse effect on the environment or human health. Microalgae are able to proliferate in a broad spectrum of adverse environmental factors like; light, salinity, pH, carbon, nitrogen, phosphorus, and temperature, etc.15 They are highly capable of capturing sunlight energy more efficiently approximately ten times higher than terrestrial plants. Microalgae multiply by capturing sunlight, CO2, H2O, and minerals.16 They generate oxygen, and biochemical energy in the form of carbohydrates, protein and lipids. These microalgae grow individually or in groups that consume carbon dioxide, nitrogen, and phosphorus, and release oxygen. Microalgae are capable to grow photoautotropically as well as heterotrophically. Photoautotrophic microalgae trap CO2 and sun energy in the form of photons for their progression and reproduction of valuable products. And also helping in the mitigation of CO2.5,11 While heterotrophic or mixotrophic microalgae take carbon and organic nutrients as an energy source from the wastewater for their growth and development while generating valuable byproducts in the form of lipids, proteins and carbohydrates, etc.17
Many researches showed the potential of microalgae for effectively eradicating heavy industrial pollution load, containing a higher amount of phosphorous, organic carbon, all forms of nitrogen, and minerals from dairy wastewater while generating biomass and value-added byproducts.18,19 The remarkable microalgal culture of “green gold” can support to create a “circular economy” that rejuvenates the environment and keep recycling the materials in nature. Hence, microalgae provide many essential ecosystem services.20 The present study emphasizes on the ability of microalgae to remediate dairy effluent while producing biomass with valuable lipids.
Poterioochromonas sp. (Chrysophyte), a group of naked unicellular biflagellate algae, are generally found in freshwater resources.21 These are having mixotrophic nutritional mode i.e., the combination of phototrophic growth via photosynthesis along with nutrient uptake via heterotrophic mode. Hence, mixotrophy seems the most thriving way of life in unfavorable conditions, to microalgae for their better survival.22 Still, microalgae remain encore urging, but the impoverished source of sustainable development.23,24 A chrysophyte isolated from dairy industry effluent and identified as P. malhamensis strain UPMC A0073 was growing swiftly in a pH range between 6.0 to 9.0 in a liquid medium and accumulated large vacuoles of lipid bodies or triacylglycerols.
This research work communicates the limited printed knowledge on chrysophytes (golden brown microalgae) as potential sources of lipid production that could be a promising bio-resource. The goal of this study was to bioremediation the dairy wastewater and the production of lipids and other biomolecules from an acid-tolerant chrysomonad, isolated from the dairy wastewater.
Dairy wastewater collection
The dairy wastewater (DWW) was collected in sterilized containers, from the Parag milk plant in Mansurpur, Muzaffarnagar (Uttar Pradesh), India, for this study and stored at 4°C until used.
Physicochemical analysis of dairy wastewater
All the suspended particles in DWW were removed by filtration process and the physicochemical characterization was carried out by using the standard protocols of water analysis i.e., APHA, 2012 (American Public Health Association)25 and metals were detected by ICP-OES. For observing the reduction in nutrients, parameters taken into consideration were, chemical oxygen demand (COD), phosphate, and nitrate before and after using microalga for treatment.
Isolation of microalgae from dairy wastewater
The 10 ml of dairy wastewater was inoculated in a conical flask capacity of 250 ml, carrying of 100 ml, Bold’s Basal Media (BBM). The successive streak plate procedure was applied for the isolation of pure microalgal sp. from mixed culture. The composition of BBM are as follows: Sodium nitrate (0.25 g L−1), Potassium dihydrogen phosphate (0.175 g L−1), Dipotassium hydrogen phosphate (0.075 g L−1), Calcium chloride (hydrated) (0.025 g L−1), Magnesium sulphate (heptahydrate) (0.075 g L−1), Sodium chloride (0.025 g L−1), and micro-elemental homogenous mixture [1 ml L−1 containing; Boric acid (2.86), Manganese chloride (1.81), Sodium molybdate dihydrate (0.390), Zink sulphate heptahydrate (0.222), and Cobalt nitrate hexahydrate (0.049), Copper sulphate (0.079) ].21,26 The pH of the broth was 7 ± 3.
Experimental design
Isolated microalgal culture, cultivated in BBM broth at 25 ± 3°C and brighten with visible light of (60 µmol m−2 s−1) with continual stirring at 100 rpm up to 15 days with 12 h light-dark cycle.27 Chloramphenicol (55 µg/ml) and ampicillin (110 µg/ml) and were mixed in to the media to avoid bacterial contamination.28 Miscellaneous microalgal species, were visible on broth, and after 15 days, repeated streaking (100 µL) was carried out on agar plate of BBM, under the same cultural conditions. After repeated streaking cycles, we obtained bacteria-free pure microalgae culture onto BBM agar plate and microscopic examination for morphological identification according to Bellinger and Sigee.29 Growth of isolated microalgal sp. was optimized by using different concentrations of dairy wastewater with growth media [25% dilution (25 ml DWW +75 ml BBM), 50% dilution (50 ml DWW+ 50 ml BBM), 75% dilution (75 ml DWW + 25 ml BBM), 100% (100 ml DWW) and Control (100 ml BBM media)]. The biomass production was determined in terms of chlorophyll, at every alternate day till the end of batch culture (15 days). The optical density was calculated at 665 nm by using Marker et al. method.30 Microalgal cells were dried for 24 hours at 60°C and DCW (dry cell weight) was measured gravimetrically. At the end of batch culture, microalgal biomass was collected, by micro centrifugation at 6000 rpm for 10 minutes. After that stable weight (DCW= dry cell weight) was obtained by drying at 60°C.
During the cultivation period, biomass productivity was determined by using the
Eq. no. (i)
Biomass productivity (BP) = (Nt _ N0)/ (tt − t0)
where, Nt is the final yield of biomass (g L−1) at the end of batch culture (15 days) (tt) and N0 is the initial yield of biomass (g L−1) at t0 (0 days).31,32
Specific growth rate (µ, d−1) was estimated by following the Eq. no. (ii)
μ = ln (Wt/ W0)/Δ t
where, Wt is the final and Wo, initial DCW (dry cell weight) of batch culture, Δ t is the change in time (days)
Cell doubling time (Td) was calculated according to equation no. (iii)33,28
(Td) =ln (2)/ μ
Nutrient removal efficiency
Approximately 100 ml of microalgae suspension was taken out from each photo-bioreactor at every 24 h. and analyzed for COD, nitrate, and phosphate removal capacity. The nutrient (COD, NO3−1, and PO4-3) removal efficiency and nutrient removal rate was calculated by the following equations no. (iv) and (v).34,35,28
Removal rate (RR) (mg/L/d) = (C0− Ct)/ (ti -to) by the equation no. (iv)
Removal percentage (RP) = [(C0 − Ct)/ C0] ×100 the equation no. (v)
Where, C0 is the initial and Ct, final concentrations at time to (at 0 day) and ti (at 15 day) respectively.
Identification of isolated microalgae
To identify the isolated microalgal strain, firstly, genomic DNA was extracted from the microalgae culture with the DNA extraction Kit (HiMedia, India), by following the instructions. For the genetic identification of the isolated microalgal strain, the fragment of the 18S rRNA gene sequences were amplified by 18SrRNA-F and 18SrRNA-R primers (18S-F: (5’GTAGTCATATGCTTGTCTC3’); 18S-R: (5’CTTCCGTCAATTCCTTTAAG3’) by using standard PCR protocols.36 For PCR amplification required, 10 µL reaction mixture consisting; Big Dye Terminator Ready Reaction Mixture: 4µL, DNA template: 1 µl, oligonucleotide primer: 2 µl; Milli Q Water: 3 µl (kit; Himedia, India). The following thermal process was performed at 95°C temperature for 3 minutes, followed by denaturation (35 cycles) at the temperature 95°C for 1 minute, annealing at 50°C, and elongation at 72°C and last followed by 3 min extension at the same temperature i.e.72°C. This was figure out on agarose gel (1.0%); in the form of a single band then PCR amplicon purified to remove contaminants by using, a PCR purification kit. After that DNA sequencing reaction was followed with 18SrRNA-Forwards and 18SrRNA-Reverse primers (18S-F: (5’GTAGTCATATGCTTGTCTC3’); 18S-R: (5’CTTCCGTCAATTCCTTTAAG3’) using Big Dye Terminator v3.1 Cycle sequencing kit with the equipment ABI 3730xl DNA Analyzer. To determine the phylogenetic relationship of isolated microalgal strain, generated Canonical sequence data of 18S rRNA gene was aligned with multiple sequence alignment using Clustal W software program. (available reference sequences retrieved at the GenBank). Phylogenetic tree was figured out by applying the Neighbour-joining method,37 which provide evolutionary and trait based homology. Jukes-Cantor model was used to determine the genetic distance between two sequences.38 Bootstrap technique, based on 1000 replicates, used for the estimation of robustness of the tree,39
Extraction and identification of lipids
After the completion of batch culture (on the 15th day), biomass was recovered by micro-centrifugation process at 6000 rpm (revolutions per minute) for 15 minutes. Then, it was washed to remove the salt with distilled water and dried at 60°C and expressed as mg l_1. Afterward, the process of lipid extraction was performed with (1:2 v/v), a mixture of chloroform/methanol according to Bligh and Dyer method.40,41 The 1:2 ratios of methanol and chloroform was added into the dry biomass and exposed in high frequency soundwaves for 10 minutes, for dissolving the intramolecular interactions. Lipid fraction was separated from the chloroform layer with the help of separating funnel. Anhydrous sodium sulphate, (2 g) was added into this fraction, for evaporating moisture. Extracted amount of lipid was transferred in the pre-weighted, round bottom flask and weight of the lipid was estimated after drying the chloroform. Microalgal lipid content demonstrated as the % composition of dry cell weight (DCW) of microalgae. Microalgal lipid productivity was calculated by applying the Eq. no. (vi).
[LP = BP× LC /100]
Where, LP = Lipid productivity (mg L-1d-1), BP = microalgal biomass productivity (mg L-1d-1), and LC = lipid content percent (w/w).
Neutral fatty acids or lipids were identified and quantified by using an instrumental technique, GC-MS (Gas chromatography Mass spectrophotometry).
Physico-chemical analysis of DWW
DWW (dairy waste water), used in this research work, was evaluated for its physico-chemical properties which represented in Table 1. The pH was acidic i.e. (5.4 ± 0.2), and the biological oxygen demand and chemical oxygen demand levels were 1278 mg L-1 and 2398 mg L−1 respectively. The total concentration of phosphate and nitrate was 88.52 and 95.60 mg L−1, respectively. Apart from these, it also contained various minerals viz., Ca, Mg, Na, K, Fe, Mn, Zn, and Ni, in trace amounts which are needed for microalgal growth and development.
Table (1):
Physico-Chemical Analysis of Dairy Effluent.
No. |
Parameters |
Dairy Effluent (Untreated) |
Unit |
|---|---|---|---|
1. |
PH |
5.4 |
– |
2. |
Colour |
Milky White |
– |
3. |
BOD |
1278 |
mg/L |
4. |
TS |
1890 |
mg/L |
5. |
TDS |
1250.5 |
mg/L |
6. |
TSS |
639.5 |
mg/L |
7. |
COD |
2398 |
mg/L |
9. |
Total Phosphate |
88.52 |
mg/L |
10. |
Total Nitrate |
95.60 |
mg/L |
11. |
Oil and Grease |
13.6 |
mg/L |
12. |
Metals |
||
13. |
Na |
64.03 |
mg/L |
14. |
K |
27.7 |
mg/L |
15. |
Mg |
10.28 |
mg/L |
16. |
Ca |
33.231 |
mg/L |
17. |
Fe |
0.161 |
mg/L |
18. |
Zn |
0.055 |
mg/L |
19. |
Mn |
0.0084 |
mg/L |
20. |
Ni |
0.0067 |
mg/L |
Identification of isolated microalgae from DWW
The microscopic examination revealed the purity and morphology of the microalgal strain. According to preliminary morphological identification, the microalga was identified as Poterioochromonas sp. (a chrysophyte). Afterward, to amplify the 18S rDNA fragment from the isolated strain, universal primers were used. These amplicons were sequenced and submitted to the NCBI database. 18S rRNA gene sequences of isolated microalgal BLAST hits sequences indicated (97.30%) similarity with Poterioochromonas malhamensis strain UPMC-A0073 Sequence id: MK834582., and the microalgal strain showed the next closest homology with Achnanthidium saprophilum Sequence id: MN592667.1 (Figure 1) Phylogenetic tree, was inferred by using 18S rRNA and internal transcribed spacer 1-5.8S internal transcribed spacer 2 (ITS1-5.8S-ITS2).
Figure 1. Phylogenetic tree, was inferred by using 18S rRNA and ITS1–5.8S-ITS2 (PNG file attached separately)
The 18S rDNA sequences exhibited the highest (97.30%) similarity with Poterioochromonas malhamensis strain UPMC-A0073).) Phylogenetic Tree was built with a software aligner system. The phylogenetic distances were calculated by using Jukes and Cantor created formula.38 All these analyses were conducted in MEGA6.39
Growth and nutrient removal rate
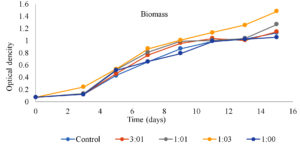
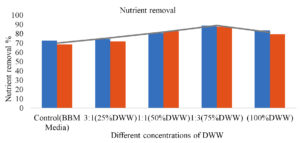
Microalgal growth and efficiency of nutrient removal directly depend on their metabolism and the nutrient’s availability in DWW. The growth of isolated microalgal sp. was observed by absorbance of light, at 650 nm by using various concentrations [25% dilution (25 ml DWW +75 ml BBM), 50% dilution (50 ml DWW +50 ml BBM), 75% dilution (75 ml DWW + 25 ml BBM), 100% (100 ml DWW) and Control (100 ml BBM media)] for enhancing the growth and removal of pollution load from DWW. Figure 2 shows the growth, in terms of biomass of isolated microalgae in different concentrations of DWW. The maximum biomass (dry cell weight) reached 1.478 g 1 in 15 days from an inoculum of 0.072 g L-1 in 75% concentration (75 ml DW+25 ml BBM media). Microalgal biomass was found to decrease slightly in DWW after 15 days. It was assumed that biomass concentration directly depends on the consumption of nutrients, in the wastewater. If the nutrients are consumed rapidly then biomass increased progressively. It was found that 75% concentration (75 ml DW+25 ml BBM media), showed better media for growth and treatment purpose than any other concentrations. However, the microalgal growth also differs according to the species. Many researchers have reported varied microalgal biomass in different conditions of DWW. Ummalyma and Sukumaran41 reported higher biomass i.e., 1.94 gL-1 when microalgae are grown with enhanced or supplemented DW. Swain et al.1 also reported that a maximum of 3.004 g L-1 DCW (dry cell weight) was achieved within 10 days in optimized media. Microalgae are able to grow in dairy effluent and generate beneficial algal biomass while removing organic and inorganic content. During the culturing time, continuous reduction of nutrients, in all the different concentrations of DWW was observed. In the present work, the highest percentage of total nitrate and phosphate removal, was 87.45% and 88.96%, respectively, observed in 75% DWW. The COD reduction was also the highest i.e., 88.84% in 75 % DWW. Some other research also showed similar observations as the present research work.34,42,1,43,44 Figure 3 represents the nutrient removal efficiency of autochthonous microalgae in different concentrations of DWW. Table 2 exemplifies the cultivation of microalgae in dairy wastewater and nutrient removal percentages (COD, NO3− and PO43−).
Table (2):
Cultivation of microalgae in dairy wastewater and nutrient removal percentages (COD, NO3− and PO43−).
Microalgae |
Biomass (g/l) |
Time (days) |
COD removal % |
Nitrate removal % |
Phosphate removal % |
Reference |
|---|---|---|---|---|---|---|
Scenedesmus sp. |
1.75 |
11 |
89.30 |
88.41 |
97.07 |
Mercedo et al.45 |
A. protothecoides |
3.30 |
10 |
65.00 |
43.00 |
77.00 |
Gramegna et al. 44 |
C.reinhardtii |
1.70 |
10 |
76.00 |
65.00 |
87.00 |
Gramegna et al.44 |
Chlorella vulgaris |
_ |
_ |
_ |
57.01 |
51.84 |
Kalaji et al.43 |
Scenedesmus quadricauda |
0.43 |
12 |
76.77 |
92.15 |
100 |
Daneshvar et al. 46 |
Scenedesmus sp. |
1.22 |
12 |
90.50 |
100 |
91.24 |
Panday et al. 28 |
Ascocloris sp. |
2.23 |
11 |
95.10 |
79.10 |
98.10 |
Kumar et al. 47 |
Poterioochromonas
Malhamensis UPMC A0073 |
1.478 |
15 |
88.84 |
87.45 |
88.96 |
In this study |
Biomass and lipid productivity
Nutrient depletion and production of biomass, simultaneously occurring in dairy waste water. Biomass and lipid productivity was greatly affected by the depletion of nutrients in the dairy wastewater. Lipid yield in the algal biomass is the crucial parameter to determine their potentiality. Table 3 summarizes the microalgal biomass characteristic i.e., biomass productivity (BP), lipid productivity (LP) and lipid content (LC%) of the isolated microalgal strain in different concentrations of DWW. Isolated microalgae showed 93.73 mg L-1 d-1 biomass productivity in 75 % DWW which was the highest BP (Biomass production) among these DWW concentrations. Isolated microalgal sp. was also able to accumulate a high amount of lipids (31.60 % w/w) in 75% concentration of DWW followed by 100% DWW (28.46% w/w), 50% DWW (26.50% w/w), control (BBM Media) (23.45% w/w), and 25% DWW (22.60% w/w). The total lipid content (LC %) (31.60% w/w) of this isolate was found to be significantly almost similar to the previously reported studies. 28,44
Table (3):
Growth characteristics of isolated microalgal sp.
Growth Characteristics |
Control (BBM Media) |
3:1 (25%DWW) |
1:1 (50%DWW) |
1:3 (75%DWW) |
1:0 100%DWW |
|---|---|---|---|---|---|
DCW (g/l) |
1.128 |
1.145 |
1.265 |
1.478 |
1.056 |
LC % (W/W) |
23.45 |
22.60 |
26.50 |
31.60 |
28.46 |
BP (mg/l/d) |
70.40 |
71.53 |
79.53 |
93.73 |
65.60 |
LP (mg/l/d) |
16.50 |
16.16 |
21.07 |
29.61 |
18.66 |
Fatty acid profiling through GCMS
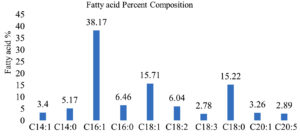
The fatty acids produced by the autochthonous microalgae in the dairy wastewater were examined by using GCMS. Major fatty acids, produced in DWW were (C16:0, C16:1, C18:1, and C18:0) found along with (C14:0, C20:1) and other fatty acids with a lower amount, which are presented in Figure 4. The amount of saturated fatty acids content of isolated microalgae was 26.83%, the unsaturated fatty acids content was 72.25%, and 0.99% others. The fatty acids of C16 to C18, were the major fatty acids while the fatty acids of C14 and C20 were presented in a minor amount. A similar profile of fatty acids with the highest proportion of hexadecanoic acid (monounsaturated fatty acid) (C16:1) followed by heptadecanoic acid (monounsaturated fatty acid) (C18:1) and heptadecanoic acid (saturated fatty acid) (C18:0) was reported for numerous microalgal sp. grown in dairy effluent.41,42 Some other previous studies also showed similar results that microalga showed high potential for lipid production when grown on dairy effluent.28,1,45,47 Table 4 shows the comparison of fatty acid lipid contents of various microalgae sp. grown in dairy waste water (DWW).
Table (4):
Comparison of fatty acids lipid content of various microalgal sp. grown in Dairy waste water.
Microalgal sp. |
Lipid % |
Lipid production (mg/l/d) |
Fatty acid composition% |
reference |
|---|---|---|---|---|
Tetraselmis sp. |
51.65 |
_ |
C16:0 20.55 C16:1 10.45 C18:0 7.72 C18:1 13.95 |
Swain et al.1 |
Scenedesmus sp. |
51.00 |
507.00 |
C16:0 18.79 C18:2 20.05 C18:3 23.79 |
Mercedo et al.45 |
A.protothecoides C.reinhardtii |
18.50
12.00 |
592.00
204.30 |
Gramegna et al.44 |
|
Scenedesmus sp. |
30.70 |
C16:0 29.23 C18:0 13.05 C18:1 46.20 C18:3 9.23 |
Panday et al. 28 |
|
Ascocloris sp.ADW007 |
34.98 |
207.00 |
C16:0 6.0 C18:0 11.5 C18:2 5.30 C18:3 9.80 C20:1 3.10 C22:6 6.60 |
Kumar et al.47 |
Chlorella sp. |
23.00 |
107.83 |
C16:0 8.44 C18:0 39.49 C18:1 27.54 C20:0 10.93 |
Choi et al.42 |
Arthrospira platensis |
30.45 |
C16:0 8.44 C18:0 39.49 C18:1 27.54 C20:0 10.93 |
Hena et al.48 |
|
Poterioochromonas Malhamensis UPMC A0073 |
31.60 |
29.61 |
C16:1 38.17 C16:0 6.46 C18:0 15.22 C18:1 15.71 C18:2 6.04 |
In this study |
The microalgal sp. isolated from the DWW was identified as Poterioochromonas malhamensis (Chrysophyte) in this study. The microalga showed high growth and lipid accumulation with different concentrations of dairy wastewater. The biomass and total lipid content percent were 1.478 g L-1 and 31.60% in optimized conditions (75% DWW) respectively, with an 88.84% reduction in COD. The fatty acid composition analysis revealed that the microalga was rich in C16:1, C18:1, and C18:0, these microalgal lipids have potential to be used in various fields such as biofuel production, nutraceuticals, pharmaceuticals, cosmetics, lubricants, etc. Conclusively, DWW contains all the required nutrients for microalgal growth and development. Hence, the utilization of DWW by microalgae cultivation is a sustainable and profitable approach. As per the information we have, this is the first report of DWW treatment utilizing the microalga Poterioochromonas malhamensis. Cultivation of which remediated the DWW, avoiding spreading contamination of freshwater sources, and produced commercially important lipids.
ACKNOWLEDGMENTS
The authors would like to thank Swami Rama Himalayan University, Dehradun, India, for providing all necessary facilities to carry out this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SG and VK conceptualized the plan of work. ND carried out the experiments and wrote the manuscript. GB and SA helped in manuscript writing. SG and VK edited the final manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Swain P, Tiwari A, Pandey A. Enhanced lipid production in Tetraselmis sp. by two stage process optimization using simulated dairy wastewater as feedstock. Biomass Bioenergy. 2020;139:105643.
Crossref - Porwal HJ, Mane AV, Velhal SG. Biodegradation of dairy effluent by using microbial isolates obtained from activated sludge. Water Resour Ind. 2015;9:1-15.
Crossref - Choi HJ. Dairy wastewater treatment using microalgae for potential biodiesel application. Environ Eng Res. 2016;21(4):393-400.
Crossref - Qadir M, Drechsel P, Cisneros BJ, et al. Global and regional potential of wastewater as a water, nutrient and energy source. Nat Resour Forum. 2020;44(1):40-51.
Crossref - Ummalyma SB, Sahoo D, Pandey A. Resource recovery through bioremediation of wastewaters and waste carbon by microalgae: a circular bioeconomy approach. Environmental Science and Pollution Research. 2021;28(42):58837-58856.
Crossref - Sharma R, Mishra A, Pant D, Malaviya P. Recent advances in microalgae-based remediation of industrial and non-industrial wastewaters with simultaneous recovery of value-added products. Bioresour Technol. 2022;344(Pt B):126129.
Crossref - Ghimpusan M, Nechifor G, Nechifor AC, Dima SO, Passeri P. Case studies on the physical-chemical parameters’ variation during three different purification approaches destined to treat wastewaters from food industry. J Environ Manage. 2017;203(Part 2):811-816.
Crossref - Ahmad T, Aadil RM, Ahmed H, et al. Treatment and utilization of dairy industrial waste: A review. Trends Food Sci Technol. 2019;88:361-372.
Crossref - Zhuang LL, Li M, Ngo HH. Non-suspended microalgae cultivation for wastewater refinery and biomass production. Bioresour Technol. 2020;308:123320.
Crossref - Singh A, Ummalyma SB, Sahoo D. Bioremediation and biomass production of microalgae cultivation in river water contaminated with pharmaceutical effluent. Bioresour Technol. 2020;307:123233.
Crossref - Hussain F, Shah SZ, Ahmad H, et al. Microalgae an ecofriendly and sustainable wastewater treatment option: Biomass application in biofuel and bio-fertilizer production. A review. Renewable and Sustainable Energy Reviews. 2021;137:110603.
Crossref - Joun J, Hong ME, Sirohi R, Sim SJ. Enhanced biomass production through a repeated sequential auto-and heterotrophic culture mode in Chlorella protothecoides. Bioresour Technol. 2021;338:125532.
Crossref - Sirohi R, Joun J, Choi HI, Gaur VK, Sim SJ. Algal glycobiotechnology: omics approaches for strain improvement. Microbial Cell Factories. 202;20(1):163.
Crossref - Kusmayadi A, Lu PH, Huang CY, Leong YK, Yen HW, Chang JS. Integrating anaerobic digestion and microalgae cultivation for dairy wastewater treatment and potential biochemicals production from the harvested microalgal biomass. Chemosphere. 2022;291(1):133057.
Crossref - Lu X, Nan F, Feng J, Lv J, Liu Q, Liu X, Xie S. Effects of different environmental factors on the growth and bioactive substance accumulation of Porphyridium purpureum. Int J Environ Res Public Health. 2020;17(7):2221.
Crossref - Kim KH, Choi IS, Kim HM, Wi SG, Bae HJ. Bioethanol production from the nutrient stress-induced microalga Chlorella vulgaris by enzymatic hydrolysis and immobilized yeast fermentation. Bioresour Technol. 2014;153:47-54.
Crossref - Ummalyma SB, Sirohi R, Udayan A, et al. Sustainable microalgal biomass production in food industry wastewater for low-cost biorefinery products: a review. Phytochem Rev. 2022:1-23.
Crossref - Nur MMA, Buma AGJ. Opportunities and challenges of microalgal cultivation on wastewater, with special focus on palm oil mill effluent and the production of high value compounds. Waste Biomass Valori. 2019;10(8):2079-2097.
Crossref - Hachicha R, Elleuch F, Ben Hlima H, et al. Biomolecules from microalgae and cyanobacteria: Applications and market survey. Applied Sciences. 2022;12(4):1924.
Crossref - Gurlek ATE, Dizdaroglu D, Yalcin D, Erkaya IA, Erdem B, & Demirel O. Landscape Research II. Livre de Lyon, 1st ED Lyon France, 2022:43-61
- Andersen RA, editor. Algal culturing techniques. Elsevier. 2005.
- Patel AK, Joun J, Sim SJ. A sustainable mixotrophic microalgae cultivation from dairy wastes for carbon credit, bioremediation and lucrative biofuels. Bioresour Technol. 2020;313:123681.
Crossref - Olabi AG, Shehata N, Sayed ET, et al. Role of microalgae in achieving sustainable development goals and circular economy. Sci Total Environ. 2022;854:158689.
Crossref - Jegathese SJP and Farid M. Microalgae as a renewable source of energy: A niche opportunity. Journal of Renewable Energy; 2014.
Crossref - Rice EW, Bridgewater L. American Public Health Association (Eds.). Standard methods for the examination of water and wastewater. 22nd ed. Washington D.C. American Public Health Association 2012.
- Morando-Grijalva CA, Vazquez-Larios AL, Alcantara-Hernandez RJ, Ortega-Clemente LA, Robledo-Narvaez PN. Isolation of a freshwater microalgae and its application for the treatment of wastewater and obtaining fatty acids from tilapia cultivation. Environ Sci Pollut Res Int. 2020;27(23):28575-28584.
Crossref - Ferrer-Alvarez YI, Ortega-Clemente LA, Perez-Legaspi IA, et al. Growth of Chlorella vulgaris and Nannochloris oculata in effluents of Tilapia farming for the production of fatty acids with potential in biofuels. Afr J Biotechnol. 2015;14(20):1710-1717.
Crossref - Pandey A, Srivastava S, Kumar S. Isolation, screening and comprehensive characterization of candidate microalgae for biofuel feedstock production and dairy effluent treatment: a sustainable approach. Bioresour Technol. 2019;293:121998.
Crossref - Bellinger EG, Sigee DC. Freshwater algae: identification, enumeration and use as bioindicators. John Wiley and Sons; 2015.
Crossref - Marker AF. The measurement of photosynthetic pigments in freshwaters and standardization of methods: conclusions and recommendations. Arch Hydrobiol Beih. 1980;14:91-106.
- Griffiths MJ, Van Hille RP, Harrison ST. The effect of nitrogen limitation on lipid productivity and cell composition in Chlorella vulgaris. Appl Microbiol Biotechnol. 2014;98(5):2345-2356.
Crossref - Brar A, Kumar M, Pareek N. Comparative appraisal of biomass production, remediation, and bioenergy generation potential of microalgae in dairy wastewater. Front Microbiol. 2019;10:678.
Crossref - Issarapayup K, Powtongsook S, Pavasant P. Flat panel airlift photobioreactors for cultivation of vegetative cells of microalga Haematococcus pluvialis. J Biotechnol. 2009;142(3-4):227-232.
Crossref - Jitha G, Madhu G. Cultivation of Oscillatoria sp. in Dairy Waste Water in Two Stage Photo Bioreactors for Biodiesel Production. Civil Engineering and Urban Planning. An International Journal (CiVEJ). 2016;3(2):87-96.
Crossref - Mishra S, Mohanty K. Comprehensive characterization of microalgal isolates and lipid-extracted biomass as zero-waste bioenergy feedstock: an integrated bioremediation and biorefinery approach. Bioresour Technol. 2019;273:177-184.
Crossref - Daniel G, Sako Y, Berland B. Phylogenetic analysis of nine species of Prorocentrum (Dinophyceae) inferred from 18S ribosomal DNA sequences, morphological comparisons, and description of Prorocentrum panamensis sp. nov. J Phycol. 1998:34(6):1055-1068.
Crossref - Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Thomas J, Cantor CR. Evolution of protein molecules. Mammalian Protein Metabolism 1969;3:21-132.
Crossref - Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911-917.
Crossref - Ummalyma SB, Sukumaran RK. Cultivation of microalgae in dairy effluent for oil production and removal of organic pollution load. Bioresour Technol. 2014;165:295-301.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Choi YK, Jang HM, Kan E. Microalgal biomass and lipid production on dairy effluent using a novel microalga, Chlorella sp. isolated from dairy wastewater. Biotechnology and Bioprocess Engineering. 2018;23(3):333-340.
Crossref - Khalaji M, Hosseini SA, Ghorbani R, et al. Treatment of dairy wastewater by microalgae Chlorella vulgaris for biofuels production. Biomass Convers Biorefin. 2021:3259-3265.
Crossref - Gramegna G, Scortica A, Scafati V, et al. Exploring the potential of microalgae in the recycling of dairy wastes. Bioresour Technol Reports. 2020;12:100604.
Crossref - Mercado I, Alvarez X, Verduga ME, Cruz A. Enhancement of biomass and lipid productivities of Scenedesmus sp. cultivated in the wastewater of the dairy industry. Processes. 2020;8(11):1458.
Crossref - Daneshvar E, Zarrinmehr MJ, Koutra E, Kornaros M, Farhadian O, Bhatnagar A. Sequential cultivation of microalgae in raw and recycled dairy wastewater: microalgal growth, wastewater treatment and biochemical composition. Bioresour Technol. 2019;27:556-564.
Crossref - Kumar AK, Sharma S, Shah E, et al. Cultivation of Ascochloris sp. ADW007-enriched microalga in raw dairy wastewater for enhanced biomass and lipid productivity. Int J Environ Sci Technol. 2019;16(2):943-954.
Crossref - Hena S, Znad H, Heong KT, Judd S. Dairy farm wastewater treatment and lipid accumulation by Arthrospira platensis. Water Research. 2018;128:267-277.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.