Gram-positive pathogens are causing many serious infections that affect humans and result in mild to severe diseases worldwide. In order to survive and initiate infection, enteric pathogens must resist the physiochemical defence factors in the human intestinal tract. One of these defence factors is bile, a potent antibacterial like compound in the intestine. Efflux pumps are the important mechanism by which bacteria resist antibacterial agents such as bile. Efflux of antimicrobial substances outside the bacterial cell is considered as a key factor for intestinal colonization and virulence of enteric pathogens. This paper will review the research conducted on efflux–mediated bile resistance in Staphylococcus aureus, Listeria monocytogenes, Enterococcus faecalis and Clostridium perfringens. These bacteria colonize in the human & animal gastrointestinal tract and they have a multiple mechanism to resist the innate defences in the gut and antibacterial activity of bile. However, bile resistance in these bacteria is not fully understood. The evidence from this review suggests that Gram-positive pathogens have the ability to active transport of bile. Further research is needed to know how these pathogens sense bile and how bile regulates its virulence factor. In general, therefore, it seems that understanding the specific mechanism of bile resistance in enteric bacteria including gram-positive pathogens may involve in the development of novel strategies to control and treatment of gastrointestinal infections.
Efflux pump, Bile salt, antimicrobial agent resistance, enteric gram-positive pathogen
Bile is a digestive secretion made by hepatocytes of the liver and enters the small intestine via the bile duct. The main functions of bile are emulsification and solubilization of lipids and as a hormone regulating food metabolism.1,2 Bile salts are the major component of bile and constitute about 50% of its organic component. It is well documented that bile salts are one of the detergents that have antibacterial action against a wide range of enteric bacteria. Enteric Gram-positive pathogens must resist the toxic action of bile salts to survive inside the intestinal tract. These toxic actions can disrupt the bacterial membrane, induce oxidative stress and DNA damage and dissociate membrane protein.2,3 Thus, these pathogens have developed different mechanisms to resist bile salts. The possible mechanisms by which bacteria resist bile include synthesis of various types of fatty acid and surface protein, reducing oxidative thiol modifications, restoring redox homeostasis, bile transport using efflux pump system & hydrolyze of bile acids using bile salt hydrolases (BSHs). However, the specific mechanisms are not fully understood yet.3-6
Bile efflux is one of the important mechanisms, whereby bacteria resist the detergent action of bile by pumping out the bile salt from their cytoplasm to the external environment. Pumping out of antibacterial agents including bile salt needs particular proteins known as efflux pumps.7 The antimicrobial and other harmful agents that affect the bacteria induce expression of the efflux pumps.8 Also, efflux pumps are more than only antibiotic-resistant factors. They even help the microorganisms to pump out the heavy metals, solvents and biocides or any harmful agents.9
After discovering the efflux pumps in the 1990s, molecular microbiology improvement led to recognise different types of efflux pumps in Gram-positive bacteria such as; methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae, Clostridium difficile, Enterococcus spp. and Listeria monocytogenes and Gram-negative bacteria like, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Stenotrophomonas maltophilia, Campylobacter jejuni, Pseudomonas aeruginosa, Vibrio cholerae and Salmonella spp.10,11
Basically, efflux pumps in bacteria are found as five groups depending on composition, source of energy and number of transmembrane spanning regions as; the resistance-nodulation-division (RND) family, the major facilitator superfamily (MFS), the ATP binding cassette (ABC) superfamily, the small multidrug resistance (SMR) family and the multidrug and toxic compound.12,8 Biologically, the resistance to the antibacterial agent means that the agent is unable to affect the target place with enough concentration that inhibits the activity of the pathogen. The resistance to tetracycline by E. coli was the first efflux pump that identified in bacteria.13,14 Also, several studies suggested that the inaction of some efflux pumps in Gram-positive pathogens results increase the sensitivity of these bacteria to bile.15 So, in this review we focused on the role of efflux pump systems on the ability of some Gram-positive pathogens such as staphylococcus aureus, Listeria monocytogenes, Enterococcus faecalis and Clostridium perfringens to resist bile and survive in the gastrointestinal tract.
Efflux pump systems in Gram-positive pathogens
The microorganisms have improved their defence mechanisms inside the host through various strategies as; inactivation of the enzymes,16,17 changing the drug targets or permeability18,19 or by efflux pump overexpression.17,11 Bacteria have either selective efflux pumps for one type of substance or nonselective to transport different classes of antibiotics.11,8 Essentially, bacterial antibiotic resistance has developed due to the exposure to the antibiotics or it could be naturally resistant to some antibiotics.20 For example, E. coli has marRAB locus that intrinsically carried by the chromosomal DNA which is responsible for E. coli resistance to some antibiotics as penicillin, cephalosporins, fluroquinolones, tetracyclines, chloramphenicol, etc.21,22
Efflux pumps are transporter proteins that are located in the plasma membrane of the bacterial cell.23 There are five efflux transporter families in prokaryotes that are responsible for antibiotic resistance. Some efflux pump transporters resist to a narrow spectrum of antibiotics as TetM in E. coli against tetracyclines or MexCD-OprJ of P. aeruginosa against fourth-generation cephalosporins. While others resist a wide range of antimicrobials such as AdeABC of A. baumanii, NorA of S. aureus and AcrAB-TolC of E. coli.24 Lately, the sixth bacterial efflux pump family has been discovered in bacteria which is called the Proteobacterial Antimicrobial Compound Efflux (PACE) superfamily.25
Since, many researches focused on multidrug resistance MDR in Gram-negative pathogens due to the complex mechanisms of MDR in these bacteria because of the double-membrane around the bacterial cell that enables the expression of a triple efflux pump system as MexA/ MexB/OprM of Pseudomonas aeruginosa or AcrA/AcrB/TolC of Enterobacteriaceae. 26-31 Here is focusing on efflux pump transporters in Gram-positive pathogens and in detail pumping of bile in S. aureus bacteria which belong to four main groups of efflux pump: major facilitator superfamily MFS, ATP-binding cassette ABC, Multidrug and Toxic Compound Extrusion MATE and small multidrug resistance SMR.32-35
MFS (major facilitator superfamily)
In Gram-positive bacteria, the MDR efflux transporters include SMR, MATE, ABC and MFS.36 MFS transporter family largely exists in prokaryotes and even in eukaryotes. Type of the transport is actively using the proton motive force (PMF) as a source of energy.10,37,38 MFS proteins are mostly 400 to 600 A.A. In length organized as 12 or 14 membrane-spanning helices and there is a large loop in location between helices six and seven.33,39-41 The MFS efflux proteins in Gram-positive bacteria that suitable examples for this family of transporters are, Nor A, NorB and MdeA in Staphylococcus aureus, LmrB, Bmr and Bmr3 in Bacillus subtilis and MefE in Streptococcus pneumoniae.10,33,39,42
SMR (small multidrug resistance)
SMR transporter family is the smallest among other efflux pump groups. It is secondary multidrug transporters and belongs to the drug/metabolite transporter (DMT) family.37 The proteins in this family basically consist of 100-120 A.A., have 4 transmembrane helices and short loops between the helices act as homodimers.33,43 These transporters utilize proton motive force to pump toxic material from the bacteria to the outside.10 SMR transporters are found in Gram-negative and Gram-positive pathogens. The EmrE isolated of E. coli is the protein that studied well and has about 110 residues.44 Also, EbrAB of Bacillus subtilis and Smr, QacG, QacH of Staphylococcus aureus are famous examples of the SMR efflux pumps in Gram-positive bacteria.45,38
MATE (Multidrug and Toxic Compound Extrusion)
This group of transporters was discovered recently and exists in Gram-negative and Gram-positive bacteria.46 MATE proteins are mostly consisting of 400 to 700 A. A. with 12 transmembrane helices and all these transporters are similar in about 40% of their sequences.37,47 MepA protein of Staphylococcus aureus is a common example to MATE transporters in Gram-positive pathogens.48 In this group of efflux pumps, different antibiotics can be pumped out of the bacterial cell as norfloxacin, chloramphenicol, ciprofloxacin, kanamycin, ampicillin and others.46 Also, two sources of energy used by MATE proteins; the PMF and the sodium ion (Na+) gradient.49
NorM protein of Vibrio parahaemolyticus is the first famous example of this group. This transporter can also extrude aminoglycosides and dyes50 In addition, MepA in S. aureus is considered as one of the well-studied MATE proteins in Gram-positive bacteria. Also, there are other transporters of MATE group were studied such as YdhE in E. coli, HmrM in Haemophilus influenzae, PmpM in P. aeruginosa and CdeA in C. difficile.8,10
ABC (ATP-binding cassette)
This group of efflux pumps is large and widely spread in prokaryotes and eukaryotes. ABC proteins are active transporters that use ATP hydrolysis in process of removing the toxic substances to the outside of the cell.43 These proteins are consisting of two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). The TMDs include six transmembrane a-helices and they are homo or heterodimers and highly hydrophobic that able to recognise the harmful substances and extrusion. While, the NBDs are hydrophilic and act as the ATPase subunit in the cytoplasm which bind and hydrolyse ATP.37,44,33,45
There is a recognition feature in ABC family that can make it different from other efflux families which is the source of energy through ATP hydrolysis. ATP binding and hydrolysis causes changes in ABC proteins structure that allows transport of materials to the outside.33 LmrA in Lactococcus lacti and Rv1217c–Rv1218c in Mycobacterium tuberculosis are ABC efflux transporters example in Gram-positive bacteria.51,39
Moreover, ABC transporters act as a triple efflux pump in Gram-negative bacteria that work with the membrane fusion protein and an outer membrane protein such as MacB of E. coli which work with the outer membrane channel (TolC) and the periplasmic adaptor (MacA).40,52 LmrA protein of L. lactis was the first ABC transporter that recognised in Gram-positive bacteria.38 Also, Sav1866 in S. aureus is the model for the ABC transporter-mediated multidrug efflux. In addition to LmrA and Sav1866 are homologous of the P-glycoprotein which are multidrug transporters in humans.53,54
Efflux pump inhibitors as new antibacterial agents
Efflux pump inhibitors EPIs are molecules that are able to block efflux pumps activity and inhibit their function to extrude the antibacterial agents out of the cell. Also, they can restore the antibiotic role against the pathogens.7,55 Depending on their origin, the EPIs are either inhibitors from plant-source or synthetic inhibitors.55 In Gram-positive bacteria, for example, efflux pump inhibitors against the NorA system of S. aureus were fully studied. Both natural and synthetic EPIs are effective against NorA in S. aureus as Genistein derived from Lupinus argenteus, Spinosan A from Dalea spinosa and Tiliroside from Herissantiatiubae as a plant-origin efflux pump inhibitor in particular.56
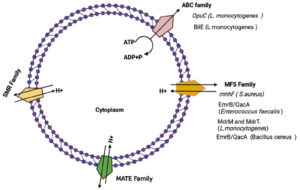
Fig. 1. Representation of different types of efflux pumps in Gram-positive bacteria and the main types of efflux pumps associated with bile resistance in some gram-positive pathogens.
It was noticed that the synthesised EPIs have toxic side effects after use in most cases. While, many natural EPIs are very effective against different groups of efflux pumps and can be used safely.55 EPIs use different mechanisms to block the efflux pumps function which results in the stop delivery of drugs or any antimicrobial factor to the outside. Thus, EPIs act as adjuvants with the antibiotics can support their role and inhibit efflux pumps transporter expression.57
Using efflux pump inhibitors EPIs appears to be the resolution to this problem or as a discovery of a new antibiotic. Also, it can assist to decrease the probability of producing a mutant generation in addition to pumping out the antibiotics.7 Moreover, EPIs mixing with antibiotics acts as a successful strategy to prevent the diseases caused by MDR pathogens via stopping the expression of efflux genes. In addition to the effect of efflux pumping the antimicrobial agent, they also decrease the concentration of the agent and may cause mutants.58
Bile efflux system in gram-positive pathogen
Enteric G-positive pathogens have multiple mechanisms to resist the detergent effect of antibacterial agents such as bile. One of the important mechanisms in the bile resistance is the efflux pump system. Several studies documented the relationship between bile resistance in gram-positive pathogen and drug efflux pumps. Sannasiddappa59 demonstration that S.aureus MnhF a secondary efflux pump involved in the resistance of this bacteria to unconjugated bile acid. He found that inactivation of Mnhf results in a significant decrease in the ability of S.aureus to resist the detergent effect of bile and the ability of the bacterium to survive in the in vitro human colonic model. Furthermore, expression of S.aureus mnhf in E.coli enhanced the resistance of the host cell to unconjugated bile acids.
EF0420 and EF1814 gene which encodes EmrB/QacA family drug resistance transporters protein in Enterococcus faecalis showed increased transcriptional activity upon exposure to 1% of bovine bile. Enterococcus faecalis EmrB/QacA efflux pump plays a role in PMF dependent extrusion of bile which is required for active extrusion of bile.60 In Listeria monocytogenes, several studies reported the role of efflux bases transporters in bile resistance. Exposure of Listeria monocytogenes to 1% porcine bile results in the induction of multidrug resistance efflux pump of the major facilitator superfamily MdrM and MdrT. L. monocytogenes MdrT mutant were found more susceptible to cholic acid and bile than wild type.61 Sleator et al58 suggest that active efflux of bile from the bacterial cell is a more effective mechanism than other mechanisms such as bile detoxification. They identify a novel bile efflux system (BilE) in Listeria monocytogenes, which is a new virulence factor in this bacterium and play a critical role in intestinal colonization. BilE belongs to ABC type MDR efflux transporter in Listeria monocytogenes and have sequence similarity to members of the betaine carnitine choline transporter (BCCT) family. The mutation in BilE gen was associated with defects in removing bile and increasing the sensitivity of the bacterium to bile.58 Sleator and colleagues59 found that OpuC, an ABC transporter has a significant role in colonizing and survival of L. monocytogenes in the small intestine. Enteric bacteria including L. monocytogenes able to resist hyperosmotic stress in the gut lumen by accumulating of active osmotic compounds such as carnitine and betaine. A recent study demonstrates that OpuC is a high-affinity carnitine transporter. Mutation in the carnitine transport system, OpuC lead to an increase of the bacterium sensitivity to bile in general, therefore it seems that L. monocytogenes OpuC has a vital role in resistant the acute toxicity of bile.62-64 Bacillus cereus a gram-positive opportunistic pathogen, which is responsible for a wide range of infections in humans such as food poisoning. It is well documented that Bacillus cereus has the ability to germinate and sporulate in the mice gut, however, little data is available about the ability of the bacterium to survive and colonize the human gut. Transcriptional response to bile shows that several gen in Bacillus cereus was unregulated. Induction of EmrB/QacA transporter family in the presence of bile refers to the role of this type of efflux pump in bile response.63 A few experimental data is available about the role of Clostridium efflux pump in bile resistance.
This assay has reviewed the role of efflux pump system in bile resistance in gram-positive enteric pathogens. The ability of enteric bacteria to resist bile is required for colonization and initiating infection. The specific mechanism by which enteric pathogen resists bile is not fully understood, however, accumulating evidence suggests that efflux pump plays a key role in bile resistance in a wide variety of enteric pathogens. These pumps transport bile outside of the bacterial cell and then decrease its harm.64,65,37 As previously mentioned, bacterial efflux pumps can be classified into six main families, including ABC, MFS, SMR, RND, MATE and PACE family. This is the first study reporting efflux-mediated bile resistance in gram-positive pathogens.37 The investigation of efflux mediated bile resistance in Staphylococcus aureus, Listeria monocytogenes, Enterococcus faecalis, Bacillus cereus and Clostridium perfringens has shown that most transporters related to bile resistance belong to ABC and MFS family. ABC family transporters need energy (ATP) to drive transport. While MFS family drives transport using the proton motive force (PMF) as a source of energy. Upregulation of gene coding efflux pump proteins upon exposure gram pathogen to a sublethal dose of bile, and increasing sensitivity to bile in pathogen lack to efflux pump (mutant) are the main methods were used in studies of bile tolerance. These studies suggest that gram-positive pathogens have the ability to active transport of bile, however Further research is needed to know how pathogenic bacteria sense bile and how bile involves in virulence factor regulation.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7(1):22-39.
Crossref - Halsey CR, Glover RC, Thomason MK, Reniere ML. The redox-responsive transcriptional regulator Rex represses fermentative metabolism and is required for Listeria monocytogenes pathogenesis. PLoS Pathogens. 2021;17(8):e1009379.
Crossref - Cremers CM, Knoefler D, Vitvitsky V, Banerjee R, Jakob U. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc Natl Acad Sci USA. 2014;111(16):E1610-E1619.
Crossref - Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179(8):2512-2518.
Crossref - Foley MH, O’Flaherty S, Allen G, et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci USA. 2021;118(6):e2017709118.
Crossref - Sharma A, Gupta VK, Pathania R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J Med Res. 2019;149(2):129-145.
Crossref - Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19(2):382-402.
Crossref - Blanco P, Hernando-Amado S, Reales-Calderon JA, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms. 2016;4(1):14.
Crossref - Schindler BD, Kaatz GW. Multidrug efflux pumps of Gram-positive bacteria. Drug Resistance Updates. 2016;27:1-13.
Crossref - Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64(2):159-204.
Crossref - Poole K. Efflux pumps as antimicrobial resistance mechanisms. Annals of Medicine. 2007;39(3):162-176.
Crossref - McMurry L, Petrucci RE, Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77(7):3974-3977.
Crossref - Zavascki AP, Goldani LZ, Li J, Nation RL. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother. 2007;60(6):1206-1215.
Crossref - Kimoto-Nira H, Kobayashi M, Nomura M, Sasaki K, Suzuki C. Bile resistance in Lactococcus lactis strains varies with cellular fatty acid composition: Analysis by using different growth media. Int J Food Microbiol. 2009;131(2-3):183-188.
Crossref - Bush K, Miller GH. Bacterial enzymatic resistance: β-lactamases and aminoglycoside-modifying enzymes. Curr Opin Microbiol. 1998;1(5):509-515.
Crossref - Sabatini S, Gosetto F, Serritella S, et al. Pyrazolo [4, 3-c][1, 2] benzothiazines 5, 5-dioxide: a promising new class of Staphylococcus aureus NorA efflux pump inhibitors. J Med Chem. 2012;55(7):3568-3572.
Crossref - Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109-1117.
Crossref - Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Bio Rev. 2003;67(4):593-656.
Crossref - Kumar S, Varela MF. Molecular mechanisms of bacterial resistance to antimicrobial agents. Chemotherapy. 2013;14:18. https://www.researchgate.net/profile/Manuel-Varela-3/publication/259621487_Molecular_mechanisms_of_bacterial_resistance
_to_antimicrobial_agents/links/0a85e532c8c7dc2790000000/Molecular-mechanisms-of-bacterial-resistance-to-antimicrobial-agents.pdf - George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155(2):531-540.
Crossref - Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41(10):2067-2075.
Crossref - Amaral L, Martins A, Spengler G, Molnar J. Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol. 2014;4:168.
Crossref - Hernando-Amado S, Blanco P, Alcalde-Rico M, et al. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resistance Updates. 2016;28:13-27.
Crossref - Hassan KA, Liu Q, Henderson PJ, Paulsen IT. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. MBio; 2015;6(1).
Crossref - Pages JM, Amaral L, Fanning S. An original deal for new molecule: reversal of efflux pump activity, a rational strategy to combat gram-negative resistant bacteria. Curr Med Chem. 2011;18(19):2969-2980.
Crossref - Martins M, Dastidar SG, Fanning S, et al. Potential role of non-antibiotics (helper compounds) in the treatment of multidrug-resistant Gram-negative infections: mechanisms for their direct and indirect activities. Int J Antimicrob Agents. 2008;31(3):198-208.
Crossref - Nikaido H, Pages JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340-363.
Crossref - Lavigne JP, Sotto A, Nicolas-Chanoine MH, et al. Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin Microbiol Infect. 2012;18(6):539-545.
Crossref - Ghisalberti D, Masi M, Pages JM, Chevalier J. Chloramphenicol and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem Biophys Res Commun. 2005;328(4):1113-1118.
Crossref - Askoura M, Mottawea W, Abujamel T, Taher I. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J Med. 2011;6:5870.
Crossref - Poole K, Lomovskaya O. Can efflux inhibitors really counter resistance? Drug Discovery Today: Therapeutic Strategies. 2006;3(2):145-152.
Crossref - Jarmula A, Oblak E, Wawrzycka D, Gutowicz J. Efflux-mediated antimicrobial multidrug resistance. Postepy Hig Med Dosw (Online). 2011;65:216-227.
Crossref - Pumbwe L, Piddock LJ. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett. 2002;206(2):185-189.
Crossref - Kehrenberg C, Schwarz S. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob Agents Chemother. 2004;48(2):615-618.
Crossref - Handzlik J, Matys A, Kiec-Kononowicz K. Recent advances in multi-drug resistance (MDR) efflux pump inhibitors of Gram-positive bacteria S. aureus. Antibiotics. 2013;2(1):28-45.
Crossref - Du D, van Veen HW, Murakami S, Pos KM, Luisi BF. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struct Biol. 2015;33:76-91.
Crossref - Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Revi. 2000;64(4):672-693.
Crossref - Borges-Walmsley MI, McKeegan KS, Walmsley AR. Structure and function of efflux pumps that confer resistance to drugs. Biochemical Journal. 2003;376(2):313-338.
Crossref - Alcalde-Rico M, Hernando-Amado S, Blanco P, Martinez JL. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol. 2016;7:1483.
Crossref - Tanabe M, Szakonyi G, Brown KA, Henderson PJ, Nield J, Byrne B. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem Biophys Res Commun. 2009;380(2):338-342.
Crossref - Markham PN, Neyfakh AA. Efflux-mediated drug resistance in Gram-positive bacteria. Curr Opin Microbiol. 2001;4(5):509-514.
Crossref - Langton KP, Henderson PJ, Herbert RB. Antibiotic resistance: multidrug efflux proteins, a common transport mechanism. Nat Prod Rep. 2005;22(4):439-451.
Crossref - Arkin IT, Russ WP, Lebendiker M, Schuldiner S. Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four-helix bundle. Biochem. 1996;35(22):7233-7238.
Crossref - Kikukawa T, Nara T, Araiso T, Miyauchi S, Kamo N. Two-component bacterial multidrug transporter, EbrAB: mutations making each component solely functional. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2006;1758(5):673-679.
Crossref - Desai H, Wong R, Pasha AK. A novel way of treating multidrug-resistant Enterococci. North American Journal of Medical Sciences. 2016;8(5):229-231.
Crossref - Wasaznik A, Grinholc M, Bielawski KP. Active efflux as the multidrug resistance mechanism. Postepy Hig Med Dosw (Online). 2009;63:123-133.
- Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27(11):587-593.
Crossref - Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2009;1794(5):763-768.
Crossref - Morita Y, Kodama K, Shiota S, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42(7):1778-1782.
Crossref - Wang K, Pei H, Huang B, et al. The expression of ABC efflux pump, Rv1217c-Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Curr Microbiol. 2013;66(3):222-226.
Crossref - Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183(19):5639-5644.
Crossref - Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69(12):1555-1623.
Crossref - Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443(7108):180-185.
Crossref - Kumar R, Pooja Patial SJ. A review on efflux pump inhibitors of gram-positive and gram-negative bacteria from plant sources. Int J Curr Microbiol Appl Sci. 2016;5(6):834-855.
Crossref - Falcao-Silva VS, Silva DA, Souza MDFV, Siqueira-Junior JP. Modulation of drug resistance in Staphylococcus aureus by a kaempferol glycoside from Herissantia tiubae (Malvaceae). Phytotherapy Research. 2009;23(10):1367-1370.
Crossref - Lomovskaya O, Warren MS, Lee A, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105-116.
Crossref - Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254-267.
Crossref - Sannasiddappa TH, Hood GA, Hanson KJ, Costabile A, Gibson GR, Clarke SR. Staphylococcus aureus MnhF mediates cholate efflux and facilitates survival under human colonic conditions. Infect Immun. 2015;83(6):2350-2357.
Crossref - Solheim M, Aakra A, Vebo H, Snipen L, Nes IF. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl Environ Microbiol. 2007;73(18):5767-5774.
Crossref - Quillin SJ, Schwartz KT, Leber JH. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol Microbiol. 2011;81(1):129-142.
Crossref - Sleator RD, Wemekamp-Kamphuis HH, Gahan CGM, Abee T, Hill C. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol. 2005;55(4):1183-1195.
Crossref - Sleator RD, Wouters J, Gahan CGM, Abee T, Hill C. Analysis of the Role of OpuC, an Osmolyte Transport System, in Salt Tolerance and Virulence Potential of Listeria monocytogenes. Appl Environ Microbiol. 2001;67(6):2692-2698.
Crossref - Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, Davies W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J Bacteriol. 2007;189(14):5302-5313.
Crossref - Sue D, Boor KJ, Wiedmann M. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiol. 2003;149(11):3247-3256.
Crossref - Gipson KS, Nickerson KP, Drenkard E, et al. The great ESKAPE: exploring the crossroads of bile and antibiotic resistance in bacterial pathogens. Infect Immun. 2020;88(10):e00865-19.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.