ISSN: 0973-7510
E-ISSN: 2581-690X
Aflatoxin B1 (AFB1) and ochratoxin A (OTA) are two of the most important of mycotoxins associated with tropical and subtropical climate as in Egypt. Therefore, this study aimed to isolated lactic acid bacteria (LAB) and Bifidobacteria from some local dairy products from Egypt and using some strains to reduce/or remove AFB1 and OTA toxins. Fifty samples, of milk and dairy products, including ten samples each, of Kareish, Damietta cheese (soft type), buffalo’s milk, yoghurt (zabady), and naturally acidified milk (Rayeb)were screened for their load of Lactobacilli, Lactococci and Bifidobacteria, using MRS agar, M17 agar and (MRS-Cys) agar, respectively. The obtained data indicated that 38, 31 and 22 isolates belonged to Lactobacillus, Lactococcus and genus Bifidobacterium, respectively. Fourteen strains isolated from LAB and Bifidobacterium were tested on AFB1 and OTA in PBs contaminated by10 ppb with three times incubation periods (6, 12 and 36 hours) at 37°C. The results indicated that all tested strains were able to reduce AFB1 at different rates ranging between 12.1 to 65.4% after incubation for 6 h. These rates increased to 78.8% and 89.9% after incubation at 37°C for 12 and 36h, respectively. The same results were with OTA, where percentages of reduction ranging from 81.4 to 80.4% were obtained when incubation with LAB cells was done at 37°C and prolonged to 36 h. According to the results, the percentage of reduction increased with the increasing time for each strain.
Lactic acid bacteria (LAB), aflatoxin B1 (AFB1), ochratoxin A (OTA)
Mycotoxins can be defined as toxic secondary metabolites that are produced by many species of fungi in the field and /or during storage. Aflatoxin B1 (AFB1) and ochratoxin A (OTA) are two of the most important mycotoxins regarding their occurrence and toxicity1. AFB1 and OTA were classified by the International Agency for Research on Cancer (IARC) as human carcinogens that belong to Group 1 and Group 2B, respectively2. Contamination by AFB1 and OTA harms human and animal health and causes big economic losses that reach billions of dollars every year. Thus, getting rid of this toxin in food and feed by using certain strategies is needed urgently. Current methods to reduce mycotoxin contamination in food and feed can be classified into physical, chemical and biological methods3,4. Many studies used the bacteria, yeasts and molds to reduce or degrade AFB1and OTA in foods and feeds5, 6. Lactic acid bacteria (LAB) are a good choice for control or reduce AFB1 and OTA from a contaminated medium, generally considered to be safe according to USFDA; some of them also have many benefits on health, which are called probiotics bacteria7. Viable and non-viable cells of LAB have an adsorbent ability to bind toxins. Where it overlaps between the toxins and the functional groups of the cell surface happens, adsorption occurs. A lot of factors like temperature, pH, time of incubation, bacterial concentration and type of strain may affect the ability of LAB to bind toxins8, 9. Therefore, this study aimed to evaluate the ability of some strains LAB, isolated from dairy products, to reduce or degrade AFB1 and OTA from liquid media at (6h, 12h and 36 h) period incubation.
Samples from dairy and dairy products
Fifty samples were collected from milk and dairy products. The collected samples included ten samples from each of buffalo’s milk, yoghurt, naturally acidified milk (Rayeb), Kareish and Domiati cheese (soft type). All samples were analyzed using three isolation media. De Man, Rogosa, Sharp agar (MRS) agar (Oxoid, UK) was used for isolation of lactobacilli, MRS medium supplemented with L-cysteine (0.5 g/liter) (MRS-Cys) agar was used for isolation of Bifidobacteria, however, M17 medium (Oxoid) agar was used for isolation of Lactococci spp.
Microbiological assessment of products
Twenty-five g/ml of each sample was aseptically transferred to conical flask 500 ml and homogenized in 225 ml of sterile tri-sodium citrate10. Ten-flods dilutions of homogenates were prepared and inoculated onto plates of MRS, MRS-Cys agar, which incubated in anaerobic condition using Co2 generated kits (Anaero-Gen. Oxoid) and incubated at 37°C for 48 h, while M17 plates were incubated in aerobic conditions at 32°C. All plates were incubated for a period of three to five days11,12.
Identification of LAB and Bifidobacterium
All the selected strains (120) isolates were activated twice in MRS-Cys, MRS and M17 broth at 37°C for 24 h. The selected isolates were observed by optical microscopy to determine their morphology and Gram staining results13. Additionally, they were tested for catalase14, oxidase, nitrate reductase, gelatinase activities, production of indol, and production of gas from glucose15. The API-50CHL identification system (BioMerieux, Lyon, France) was used for Lactobacilii and Bifidobacterium, whereas the API- 20 STREP identification system (Bio-Merieux, Lyon, France) was used for Lactococci.
Preparation of culture cells
The strains of lactic acid bacteria were activated overnight (16 h) in 100 ml MRS, MRS-Cys and M17 broth. A cell free solution was obtained by centrifugation of culture at 5000 rpm at 4°C for 20 min.
Preparation of AFB1 and OTA spiked solution
AFB1 and OTA standards were purchased from Sigma, chemical Co. (St. Louis, MO, U.S.A). Stock solutions and standards were prepared and assayed according to AOAC Method 971.22, 2005)16. Standards separately were added by 10 ppb to 100 mL phosphate buffered saline (PBS).17All solvents were of HPLC grade. The water was double distilled with Millipore water purification system (Bedford, M A, USA).
AFB1 and OTA binding assay in vitro
One hundred milligrams of bacterial cells were added to100 ml of PBs contaminated by 10 ppb of AFB1 and OTA both are separate from each other. Samples were incubated at 37°C for 6, 12 and 36h for testability of strains for binding of toxins. Then in each incubation period; filtration for discarding bacterial cells was done through filtration then extraction and determination of AFB1 or OTA in PBs (100 mL) with three replicates for each incubation time.
Extraction and determination of AFB1using HPLC
AFB1 was extracted and cleanup using immunoaffinity column (IAC) Aflatest ®-p affinity to obtained the dry film, then determination AFB1 in samples after derivatives process by adding 100μl of trifluoroacetic acid (TFA) to samples and mixed well for 30 s and the mixture stands for15 min, finally measurement by HPLC)17
Extraction and determination of OTA using HPLC
OTA was extracted and cleaned up using the method described in AOAC, (2007)18 using the IAC (Ochra Test ®-p affinity column), then concentrations of OTA were determined by High- Performance Liquid Chromatography (HPLC) system19.
Statistical analyses
All data were statistically analyzed using the General Linear Model procedure of the SPSS ver. 18 (IBM Corp, NY). The significance of the differences among treatment groups was determined by Waller–Duncan k-ratio. All statements of significance were based on the probability of P- value ≤0.05 was considered to be statistically significant.
Isolation of lactic acid bacteria and Bifidobacterium
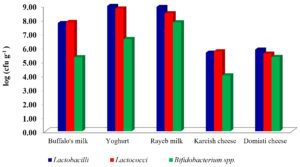
A total of fifty samples of milk and dairy products were investigated for their load of LAB and Bifidobacterium spp. This included 10 samples, each of buffalo’s and Rayeb milk, yoghurt, Kareish and Domiati cheese. The results showed in (Fig. 1) as an average log of a colony-forming unit (cfu) of LAB and Bifidobacterium spp.
Yoghurt was the highest product in log count among all other products, for example the count of Lactobacilli was 8.97 cfu g-1, and 8.79 cfu g-1 for Lactococci, whereas kareish cheese was the lowest product in log count among all other products. For example, the count of Lactobacilli was 5.62cfu g-1 and 4.00 for Bifidobacterium. The same point of view was adapted by El-Kholy et al., (2016)10 who reported that the highest counts of L. reuteri were recorded for functional Tallaga cheese made with 3, 5 and 10% of mushroom. However, the results of this study were similar to Hussein et al., (2017)20, who reported that, counts of Lb. bulgaricus and S. thermophilus increased until day 3 in all yoghurt treatments (p>0.05), then declined thereafter. The obtained data in Table (1) showed that 250 isolates were picked up from the specific selective media of Lactobacilli, Lactococci and Bifidobacterium. All the selected isolates of LAB and Bifidobacterium were Catalase-negative and Gram-positive. Ninety-two isolates were spherical; however, the other 158 isolates were rod shaped. These isolates were taken to be identified according to their morphological, cultural characteristics according to Sneath et al., (2009)21and 50CHL API- 20STREP identification system (BioMerieux).
Table (1):
LAB isolated from some dairy and dairy products samples.
| Products | Lactobacilli | Lactococci | Bifidobacterium spp. | |||
|---|---|---|---|---|---|---|
| Isolates | Strain | Isolates | Strain | Isolates | Strain | |
| buffalo’s milk | 14 | 6 | 15 | 4 | 13 | 4 |
| Yoghurt | 22 | 9 | 21 | 8 | 17 | 5 |
| Rayeb milk | 24 | 10 | 25 | 7 | 21 | 6 |
| Kareishcheese | 13 | 6 | 14 | 5 | 8 | 3 |
| Domiati cheese | 16 | 7 | 17 | 7 | 10 | 4 |
| Total | 89 | 38 | 92 | 31 | 69 | 22 |
Among the identified 91 LAB and Bifidobacterium strains, 31 isolates belong to genus Lactococcus, while the other 38 isolates belong to genus Lactobacillus and 22 isolates belong to genus Bifidobacterium. The species which represent the total cocci were identified as (9 strains) Lactococcuslactis sub sp. Lactis, (4 strains) Leuconostoc mesenteroides, (11 strains) Lactococcus cremoris and (7 strains) Streptococcus thermophilus. On the other hand, the identified strains belonging to genus Lactobacillus,8 strains of them belonged to Lactobacillus acidophilus, 3 strains of them belonged to Lactobacillus caseisub sp. rhamnosus, 5 strains of them belonged to Lactobacillus plantarum, 4 strains of them belonged to Lactobacillus helveticus, 3 strains of them belonged to Lactobacillus gasseri, 4 strains of them belonged to Lactobacillus reuteri, 6 strains of them belonged to Lactobacillus delbrueckii sub sp. bulgaricus and 5 strains of them belonged to Lactobacillus casei sub sp. casei. Furthermore, the identified strains belonging to genus Bifidobacterium 9 strains of them belonged to Bifidobacterium bifidum and 13 strains of them belonged to Bifidobacterium breve.
Evaluation of efficacy of some LAB strains to reduce AFB1
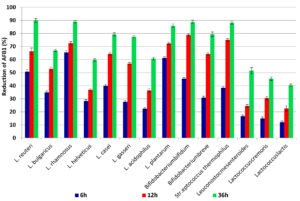
The results indicated that using L. rhamnosus gave the highest reduction for AFB1 after incubation at 37°C for 6, 12 and 36 h, where 65.4%, 72.6% and 89.0%, respectively. When using L. plantarum for removing or degradation of AFB1, the increase in removal with increased time of incubation was 61.3%, 72.4 and 85.9% after 6, 12 and 36h, respectively. While Bifidobacterium bifidum and Streptococcus thermophilus reduced AFB1 88.8 and 88.3% after 36h of incubation time, respectively. Generally, the minimum reduction percentages of AFB1 were 12.1, 22.6 and 40.4%after incubation for 6h, 12h and 36h, respectively with Lactococcus lactis. ANOVA analysis of the effect of incubation time and type of LAB strains on the content of AFB1 in liquid media is shown in (Table 2), which shows that there is a significant effect due to the incubation period as well as LAB strain. The obtained data shown in (Fig. 2) reflected that bind of AFB1 depends heavily on incubation time, where the highest binding occurred after incubation for36h using L. reuteri and L. rhamnosus89.9 and 89.0%, respectively, moreover Bifidobacterium bifidum and Stre. thermophilus removed 88.8% and 88.3% from AFB1 at the same time, respectively. Many previous studies reported that the binding of AFB1 by LAB depends on several factors such as incubation time, concentration of toxin, bacterial counts, temperature and pH. L. rhamnosus and L. acidophilius successfully got rid of nearly 80 and 60% of AFB1; respectively in PBS. They also noticed a rapid toxin binding average (80%) in 1 h by LAB22. Peltonen et al., (2001)23, reported that L. rhamnosus could successfully bind>50% of AFB1 content in PBs at 37°C within 24 h. On the other hand, Kankaanpaa et al., (2000)24, noticed that the ability of AFB1to bind in vitro and in vivo depended on LAB strain. Though the complex formed between AFB1 and bacteria was proved to be stable, the mechanism of binding between AFB1 and LAB was not illustrated yet. It was thought that AFB1 binds to cell wall components, including both polysaccharides and peptidoglycans. It was identified before that both hydrogen bonds and Van der Waals interactions were involved in AFB1 binding25-27. AFB1 binds by the LAB cell wall during processes namely adsorption and desorption, primarily, the AFB1 molecule is trapped inside the single helix of the (1 ➔ 3)-β-D-glucan chain. Then, the AFB1 molecule is covered by the branched (1➔6)-β-D-glucan chain, where it is preserved inside the helix. It also showed some involvement with both electrostatic interactions and hydrogen bonds between AFB1 molecules and the bacterial cell wall. Furthermore, AFB1 can be bound to the bacterial cell wall through weak non-covalent interactions such as associating with hydrophobic pockets on the bacterial surface. The differences in AFB1 binding by the strains are probably due to the different bacterial cell wall and cell envelope structure28, 29.
Table (2):
ANOVA of the effect of different incubation time and type of LAB strains on reductionAFB1.
| Source | SS | df | MS | F | P |
|---|---|---|---|---|---|
| Intercept | 360767.631 | 1 | 360767.6 | 59956.35 | 0.000000 |
| LAB | 35011.1 | 13 | 2693.161 | 447.579 | 0.000000 |
| Time | 28302.06 | 2 | 14151.03 | 2351.774 | 0.000000 |
| LAB*time | 2778.899 | 26 | 106.881 | 17.763 | 0.000000 |
| Error | 505.442 | 84 | 6.017 | ||
| Total | 427365.1 | 126 | |||
SS: Sum of Squares, df: degree of freedom, MS: mean square, P: probability at confidence 0.95
Fig. 2. The percentages of reduction AFB1 after incubation for 6h, 12h and 36h at 37°C with LAB cells.
Impact of LAB on the content of OTA in contaminated PBs medium
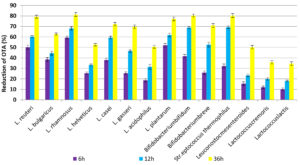
The obtained results indicated that the highest reduction percentage of OTA after incubation for 6h was 59.3, 52.0 and 50.2% after incubation with cells from L. rhamnosus, L. plantarum and L. reuteri, respectively. Besides, the lowest reduction was 10.5, 12.1 and 15.5% in samples incubated with Lactococcuslactis, Lactococcus cremoris and Leuconostoc mesenteroides, respectively. Incubation with L. rhamnosus for 12 and 36h removed 68.1 and 81.4% from OTA, respectively. On the other hand Str. thermophilus and Bifidobacterium bifidum cells removed 80.3 and 80.4% from OTA after incubation for 36h at 37°C. The ANOVA analysis in (Table 3) showed significant differences between reduction percentages of OTA after incubation with different LAB cells as well as significant differences in incubation time. These results indicated that the incubation time was one of the important factors for the efficiency of LAB cells to bind OTA in PBs. Increasing the incubation time to 12 and 36h led to increasing the reduction of OTA in contaminated PBs as shown in Fig. 3. These results are consistent with many studies conducted in this field29, where Małgorzata, (2014 )30 found that in the case of live bacterial cells, OTA decrease ranged from 16.9% to 35% in MRS medium and from 14.8% to 26.4% in PBS after 12 h, while thermally inactivated bacterial biomass gave the higher binding to AFB1 (46.2% to 59.8%). Furthermore, Del Prete et al. (2007)31 investigated the fifteen strains belonging to five relevant oenological LAB species that were grown in broth synthetic culture medium spiked with OTA. The portion of OTA removed during the bacterial growth was 8 to 28%. The OTA was removed from the supernatants between 31 to 57% by the bacterial pellet. The most effective mechanism to remove OTA is binding with the bacterial cell wall, where it contained peptidoglycan matrices, polysaccharides, as well as teichoic and lipoteichoic acid, and a protein S- layer. However, binding is based on the adsorption capacity of toxins to the cells and not on enzyme activity. This is where peptidoglycan and exopolysaccharides play an important role. LAB which is thermally inactivated gives higher removal capacity may due to changes on the cell surface. Another proposed hypothesis, namely binding of toxin by bacterial cells is also influenced by electron donor-acceptor and Lewis acid-base interactions, this may due to that the cells of LAB consider as strong electron donors and weak electron acceptors, which is supported by the hydrophilic nature of their surface 32,33.
Table (3):
ANOVA for effect of incubation time and type of LAB strains on reduction OTA.
| Source | SS | df | MS | F | P |
|---|---|---|---|---|---|
| Intercept | 284572.571 | 1 | 284572.571 | 8399.328 | 0.000000 |
| LAB | 31173.733 | 13 | 2397.979 | 70.77779 | 0.000000 |
| Time | 17536.946 | 2 | 8768.473 | 258.8067 | 0.000000 |
| LAB*time | 5172.956 | 26 | 198.960 | 5.872418 | 0.000000 |
| Error | 2845.953 | 84 | 33.880 | ||
| Total | 341302.160 | 126 | |||
SS: Sum of Squares, df: degree of freedom, MS: mean square, P: probability at confidence 0.95.
Incubation of LAB cells with PBs contaminated with AFB1 and OTA might reduce the toxin contamination. The obtained results indicated that all strains were able to reduce AFB1 and OTA but at different rates which ranged between 10 to 50% with incubation for 6h and increased to 75 % after incubation for 12h. Moreover, increasing the incubation time with LAB cells to 36h led to removing 80 and more than 88% from AFB1 and OTA in contaminated PBs.
ACKNOWLEDGMENTS
We would like to express our heartfelt thanks to National Research Centre and central mycotoxins Lab as well as Department of Dairy Science for providing the necessary facilities during this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by the National Research Centre,Egypt (Project No. 11040205).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Siciliano I, Dal Bell, B, Zeppa G, Spadaro D, Gullino ML. Static hot air and infrared rays roasting are efficient methods for aflatoxin decontamination on hazelnuts. Toxins. 2017;9(2):72.
Crossref - (IARC) International Agency for Research on Cancer. Some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans. 1993:56.
- Alshannaq A, Yu JH. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int J Environ Res Public Health. 2017;14(6):632.
Crossref - Blanksona GK, Mills-Robertsonb FC, Ofosu IW. Survey of occurrence levels of aflatoxins in selected locally processed cereal based foods for human consumption from Ghana. Food Control. 2019;95:170-175.
Crossref - Ji C, Fan Y, Zhao L. Review on biological degradation of mycotoxins. Anim Nutr. 2016;2(3): 127-133.
Crossref - Fouad MT, El-Desouky TA. Anti-toxigenic effect of lactic acid bacteria against aspergillus spp isolated from wheat grains. The Open Microbiology Journal. 2020;14:252-259.
Crossref - Fuchs S, Sontag G, Stidl R, Ehrlich V, Kundi M, Knasmuller S. Detoxification of patulin and ochratoxin A, two abundant mycotoxins, by lactic acid bacteria. Food Chem. Toxicol. 2008:46(4):1398-1407.
Crossref - Sezer CA, Guven NB, Oral NB, Vatansever L. Detoxification of aflatoxin B1 by bacteriocins and bacteriocinogenic lactic acid bacteria. Turk J Vet Anim Sci. 2013;37:594-601.
Crossref - Salim A-B, Naga MEM, Fouad MT. Behavior of ochratoxin A (OTA) during bread making enrichment with some natural antioxidants. RJPBCS. 2016;7(4):689-699.
- El-Kholy W, El-Khalek ABA, Mohamed SHS, Fouad MT, Kassem JM. Tallaga Cheese as a New Functional Dairy Product. Am J Food Technol. 2016;11(5):182-192.
Crossref - Abosereh NA, El Ghani SA, Gomaa RS, Fouad MT. Molecular identification of potential probiotic lactic acid bacteria strains isolated from Egyptian traditional fermented dairy products. Biotechnology. 2016;15(1-2):35-43.
Crossref - El-Shenawy M, Fouad MT, Hassan L, Seleet F, El-Aziz MA. A probiotic beverage made from Tiger-nut extract and milk permeate. Pak J Biol Sci. 2019;22(4):180-187.
Crossref - Brock TD, Madigan MT, Martink JM, Parker J. Cell Biology of Microorganismis. Prentice Hall Int. Ed. Toronto, Canda. 1994;43.
- Herrero M, Mago B, Gonzalez B, Suarez JE. Evaluation of technologically important traits in lactic acid bacteria isolated from spontaneous fermentations. J Appl Bacteriol. 1996;81(5):565-570.
Crossref - Kandler O, Weiss N. Genus Lactobacllus. In: Bergey’s Manual of Systematic Bacteriology. Smeath PHA, NSMair, ME Sharpe (Eds): Williams and Wilkins, Baltimore, M. D. USA. 1986;2:1209.
- AOAC. Association of Official Analytical Chemists. Official methods of analysis 18th. Washington D.C., USA. 2005.
- El-Desouky TA, Ammar HAM. Honey mediated silver nanoparticles and their inhibitory effect on aflatoxins and ochratoxin A. Journal of Applied Pharmaceutical Science. 2016;6(6):83-90.
Crossref - AOAC. Official Methods of Analysis of AOAC International AOAC, Arlington, Va, USA. 2007.
- May MA Amer, Amal MH Abdel-Haleem, Tarek A. El-Desouky. Production of some chemically modified adsorbents from some field crops by- products to reduce ochratoxin A. International Journal of Food Science and Biotechnology. 2018;3(2):46-53.
Crossref - Hussein AS, Fouad MT, El-Aziz MA, Ashour NEl-N, Mostafa EAM. Evaluation of Physico-chemical Properties of Some Date Varieties and Yoghurt Made with its Syrups. J Biol Sci. 2017;17(5):213-221.
Crossref - Sneath PHA, Mair NS, Sharpe M, Elizabeth, Holt JG. Bergy’s manual of systematic bacteriology. (Eds.) Pub.Williams and Wilkins. 2009:2605.
- El-Nezami H, Kankaanpaa P, Salminen S, Ahokas J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem Toxicol. 1998;36(4):321-326.
Crossref - Peltonen K, El-Nezami H, Haskard C, Ahokas J, Salminen S. Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J Dairy Sci. 2001;84(10):2152-2156.
Crossref - Kankaanpaa P, Tuomola E, El-Nezami H, Ahokas J, Salminen SJ. Binding of aflatoxin B1 alters the adhesion properties of Lactobacillus rhamnosus strain GG in a Caco-2 model. J Food Prot. 2000;63(3):412-414.
Crossref - Shetty PH, Jespersen L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006;17(2):48-55.
Crossref - Haskard CA, El-Nezami HS, Kankaanpaa PE, Salminen S, Ahokas JT. Surface binding of aflatoxin B1 by lactic acid bacteria. Appl Environ Microbiol. 2001;67(7):3086-3091.
Crossref - Bovo F, Corassin CH, Rosim RE, de Oliveira CA. Efficiency of lactic acid bacteria strains for decontamination of aflatoxin M1 in phosphate buffer saline solution and in skimmed milk. Food Bioprocess Technol. 2013;6:2230-2234.
Crossref - Yiannikouris A, Andre G, Poughon L, et al. Chemical and conformational study of the interactions involved in mycotoxin complexation with β-D-glucans. Biomacromolecules. 2006;7(4):1147-1155.
Crossref - Hernandez-Mendoza A, Garia HS, Steele L. Screening of Lactobacillus casei strains for their ability to bind aflatoxin B1. Food Chem Toxicol. 2009;47(6):1064-1068.
Crossref - Piotrowska M. The adsorption of ohratoxin A by Lactobacillus Species. Toxins. 2014;6:2826-2839.
Crossref - Del Prete V, Rodriguez H, Carrascosa AV, de las Rivas B, Garcia-Moruno E, Munoz R. In vitro removal of ochratoxin a by wine lactic acid bacteria. J Food Prot. 2007;70(9):2155-2160.
Crossref - Var I, Kabak B, Brandon EFA, Blokland MH. Effects of probiotic bacteria on the bioaccessibility of aflatoxin B1 and ochratoxin A using an in vitro digestion model under fed conditions. Environ Sci Health B-Pestic. 2009;44(5):472-480.
Crossref - Perczak A, Golinski P, Bryla M, Waskiewicz A. The efficiency of lactic acid bacteria against pathogenic fungi and mycotoxins. Arh Hig Rada Toksikol. 2018;69:32-45.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.