ISSN: 0973-7510
E-ISSN: 2581-690X
Growth differentiation factor 9 (GDF9) is play a critical role in ovarian follicular development and ovulation rate. The present research was performed to investigate the correlation between single nucleotide polymorphism (SNP) of GDF9gene and reproductive performance such as fertility, sterility and follicles condition in Awassi breed. Forty pairs of ovaries from total 40 slaughtered Iraqi Awassi ewes were used in this study. Ovaries (20 n) were picked up from sterile ewes and another 20 gathered from fertile ewes. Genomic DNA was extracted from each ovary of the two groups and PCR-sequencing was applied to detect GDF9 gene polymorphism. Follicles and oocytes evaluation of samples (40 n) were done in each ovary and then compared with the genotyping. Furthermore, the histological and microscopic evaluations were performed for 40 ovarian tissues of the two groups. The sequence analysis revealed that there exist three SNPs in exon I; T(114)C, G(129)R and G(199)A, the 1st two were silent mutation and the last mutation was missense responsible for a glutamic acid →lysine substitution at position 67. In sterile ewes, the current study appeared higher significant increased (P<0.01) in GG genotypes at G(129)R locus and AA at locus G(199)A and significant increased (P<0.05) in CC genotype in T(114)C locus. Likewise, GA in G(129)R locus, GG genotypes in locus G(199)A reported higher significant increment (P<0.01), TT genotype in locus T(114)C recorded significantly increased (P<0.05) in fertile ewes. The GA and GG genotypes were recorded significant increased (P<0.05) in percentage of follicles (4-8mm) and oocytes number for G(129)R and G(199)A locus as compared with wild GG and mutant AA respectively, while non-significant differences were recorded between CC and TT genotype at T(114)C locus. The histological examination revealed a hypoplasia in ovarian tissue of sterile ewes accompanied with fibrous connective tissue invasion with follicles degeneration, whilstin fertile ewes;the ovarian tissues was normal with a presence of numerous corpus albicans and degenerative corpus leutium. This study concluded that the homozygote mutation is reported to be in fertile and minimizing number of follicles and oocyte (sterility), whilst the heterozygote mutation shows strong association with fertile Awassi ewes.
Homozygote, Infertility, Heterozygote, Fertility.
Growth differentiation factor (GDF) 9 is a β superfamily transforming growth factor (Elvin et al. 2000).The GDF9 secreted particularly from oocyte through folliculogenesis, it works synergistically with bone morphogenetic protein 15(BMP15) to enhance proliferationof granulosa cells, increment in inhibinproduction and suppressing progesterone secretioninduced by FSH (McNatty et al. 2005). In addition to that, GDF9 stimulate follicular growth, granulosa cells luteinization and maintain normal follicular environment in sheep (Juengel et al. 2002), maintains normal fertility in female (De Castro et al. 2016). In sheep, GDF9 is express exclusively in oocyte (Bodensteiner et al. 1999) and act paracrine to control several enzymes of granulosa cells (Elvinet al. 1996), these events are crucial for ovulation, fertilization, and successful reproduction. In the sheep, Fabre et al., (2006) notice the strong action of GDF-9 on diminishing both basal and FSH-stimulated progesterone generation via granulosa cells of small antral follicles.
Block follicular progression and complete infertility will occur in GDF9 gene deletion because ovary are occupied by abnormal follicle with an only single layer of granulosa cells that are arranged asymmetrically, in addition, theca cell layer is likewise prominently missing in all primary one-layer follicle (Dong et al. 1996).
The ovine GDF9 gene map in chromosome five (Sadighi et al. 2002). The GDF9gene spans about 2.5 kilo bases (kb), it contains two exons and single 1126 basepairs(bp) intron that intermediate between the two exons, exon I spans 397 bp that encodes 1-134 amino acids, while exon II spans 968 bp that encodes 135-456 amino acids (Bodensteiner et al. 1999).
Awassi sheep is representing more than half percent of total Iraq’s sheep (Al-Barzinji et al. 2013). Generally, about 9% of Iraqi Awassi ewes lambing twice per year (Lafi et al. 2009). The distinguishing of DNA markers can prompt the hereditary betterment and enhance selection to hasten the reproduction regimes(Asadpour et al. 2012).
Sheep litter size (ovulation rate) is an important fertility characteristic that have an economic value (Notter, 2008). Conversely, the sterility due to small and undeveloped ovaries caused by point mutations of some genes (Davis, 2005). For that reason, the enhancement of sheep fertility traits via selection is a major goal of researchers (Kumm, 2008).
The GDF9 gene has a vital role in ovulation rate enhancement and improves litter size in heterozygous of female sheep (Souza, 2014). Many mutations in GDF9 sheep gene were determined in various sheep breed, these mutations are significantly associated with infertility or with ovulation rate improvement (Hanrahan et al. 2014).
The available information regarding this aspect in Iraq is insufficient, therefore, the current study conducted to evaluate the combination between SNPs of GDF9 sheep gene with reproductive sterility and fertility in Awassi breed.
Experimental animals
Forty slaughtered Awassi ewes were used to recovered ovaries from Al-shoala abattoir/ Baghdad. Forty pairs of ovaries were picked up and transferred to the laboratory of Biotechnology Research Center-Al-Nahrian University/Baghdad within 2-3 hour. Samples that collected from fertile ewes were (20 n) and from sterile ewes (20 n) depending on case history of normal and abnormal reproductive efficiency and depend on grossly evaluation of the ovaries and genitalia condition after slaughtering. infertile with poorly observed follicles, which were predicted sterile ewes and the rest collected from normal cyclic ewes with presence follicles, corpus albicans, and some sample have corpus leutium (fertile).
Follicle and oocytes evaluation
Total 70 follicles with diameter (4-8 mm) were measured grossly with a micrometer (Mitutoyo/Japan) immediately after transferred ovaries (40 n) to a petri dish containing normal saline. Cumulus oocyte complexes (COCs) (n 88) were collected by aspiration from follicles (4–8 mm) and by slicing of ovaries, then, examined under microscope to evaluate the quality and numbers of oocyte that have been recovered.
DNA extraction and gene amplification
Genomic DNA was isolated directly from ovarian tissue of the fertile and sterile groups by G-spin Kit (INtRON/ Korea) according to manufacturer’s kit protocol.
Two primers were utilized to amplify Exon I of ovine GDF9; (forward) 5-GAAGACTGGTATGGGGAAATG-3 and (reverse): 5-CCAATCTGCTCCTACACA CCT-3. The reaction program was 35 cycles; initial denaturation 94°C for 5 min, denaturation 94°C for 45 s, annealing 58°C for 40 s and extension 72°C for 1 min, then followed by final extension 72°C for 10 min (Hafezian, 2011).
Sequencing and genotyping
The PCR products (amplicon) were sequencing successfully via Macrogen Corporation/ Korea (Sanger sequencing method). Homology search was conducted by using BLAST option, which is available online in NCBI By using BioEditprogram and NCBI, the SNPs were determined.
Histology and microscopic evaluation
First, ovaries (20 n equally from fertile and sterile ewes group) were used after genotyping. Each ovarian sample was stabilized in neutral buffered formalin (10%) (BDH/ England) after dissected. Next, the samples were transmitted into ethanol (70%) (BDH/ England), dehydration and embedding of samples in paraffin wax were done. After that, samples were cut into several sections (5 mM). After that, sections were floated into slide and left several hours to dry at 50°C, then dewaxed and rehydrated in a graded series of ethanol solutions. Finally, morphological inspection was done after hematoxylin and eosin staining (vector/USA) of the sections according to (Elvin et al. 1999; Nicol et al. 2009).
Statistical Analyses
The Statistical Analysis System- SAS (2012) program was applied to monitor the effect of factors in parameters of present study; Chi-square test was utilized for significant comparison between percentages, while T test was used to significant compare between means.
PCR amplification and sequencing
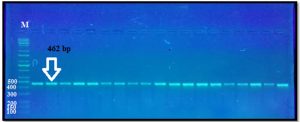
The PCR amplification of exon I ovine GDF9gene for Awassi ewes appeared uniform fragments with size 462 bp when electrophoresed in 1% agarose gel (Fig. 1).
Fig. 1. The PCR product of exon I GDF-9 gene. M = DNA ladder 100–10000 bp.
Lane 1–19 = s PCR samples.
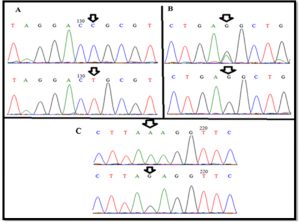
The sequencing revealed three SNPs in GDF9 gene(exon I)in chromosome 5. These SNPs; T(114)C, G(129)R and G(199)A when compared to GDF9 gene of Norwegian white sheep breed, Sequence ID: HE866499.1. Based on our sequence analysis, the SNPs identified in three different sequences, the first and third were silent mutation and the second was missense mutation, while other sample not showed any variation when compared to same breed (Fig. 2) (Table 1).
Table (1):
Type of substitution of exon I GDF-9 gene in Awassi ewes.
Location of SNP (exon I ) |
Nucleotide change |
Amino acid change & number |
Predicted effect |
Type of mutation |
|
|---|---|---|---|---|---|
1 |
T(114)C |
ACT > ACC |
Threonine > threonine (38) |
Transition |
Silent Mutation |
2 |
G(129)R |
GAG > GAG& GAA |
glutamic acid > glutamic acid (43) |
Transition |
Silent Mutation |
3 |
G(199)A |
GAG > AAG |
glutamic acid > lysine (67) |
Transition |
Missense |
The genotypic frequencies, allele distribution and sequence polymorphisms of exon I Ovisaries GDF9 gene in Awassi ewes showed in (Table 2).
Table (2):
Genotypesand alleles structure frequencies of exon I of GDF-9 gene locus for Awassi ewes.
| Locus | Genotype | Observed genotypes | Genotypic frequency % | Allele frequency | Chi square | ||
|---|---|---|---|---|---|---|---|
| 1 | T(114)C Homozygote |
TT | 18 | 45.00 | T Allele | 0.45 | 4.392 * |
| CC | 22 | 55.00 | C Allele | 0.55 | |||
| 2 | G(129)R Heterozygote |
GG | 13 | 32.50 | A Allele | 0.33 | 10.053 ** |
| GA | 27 | 67.50 | G Allele | 0.66 | |||
| 3 | G(199)A Homozygote |
GG | 16 | 40.00 | G Allele | 0.4 | 7.250 ** |
| AA | 24 | 60.00 | A Allele | 0.6 | |||
* (P<0.05), ** (P<0.01).
Association between genotypes, sterility and fertility
According to table 3, the analysis of samples that collected from sterile ewes showed that homozygote G and A alleles (GG & AA genotypes) were highly significant increased (P<0.01) than heterozygote G allele (GA genotype) in locus G(129)R and G allele (GG genotype) of G(199)A locus respectively. Additionally, in locus T(114)C; the T allele was significantly lower (P<0.05) than C allele. The results of fertile ewes showed that heterozygote G allele (GA genotype) of locus G(129)R and A allele of G(199)A locus were highly significant increased (P<0.01) than homozygote G allele and A allele of G(199)A respectively. While, the C allele was lesssignificantly (P<0.05) than T allele in locus T(114)C.
Table (3):
The correlation between sterile (20 n) and fertile ewes (20 n) with genotypes.
| parameters | T(114)C | G(129)R | G(199)A | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | Sterile | TT | CC | GG | GA | GG | AA | |
| 9 (55%) | 11 (45%) | 13 (65%) | 7 (35%) | 8 (40%) | 12 (60%) | |||
| Chi square | 4.392 * | 9.783 ** | 7.250 ** | |||||
| 2 | Fertile | 11 (55%) | 9 (45%) | 8 (40%) | 12 (60%) | 14 (70%) | 6 (30%) | |
| Chi square | 4.392 * | 7.250 ** | 11.250 ** | |||||
* (P<0.05), ** (P<0.01).
Association between genotypes and assessment of follicles and oocytes
The follicles percentage that obtained from ovarian samples that possess GA and GG genotypes were significantly higher (P<0.05) as compared withGG and mutant AA genotypes in G(129)R and G(199)A locus respectively, while non-significant differences were observed between TT and CC genotypes of T(114)C locus (Table 4).
A significant differences (P<0.05) were showed between (GG and GA genotypes) and between (GG and AA genotypes) in percentage of oocytes that completely and partially enclosed with cumulus cells (CCs) in G(129)R and G(199)A locus respectively, but in T(114)C locus, non-significant differences were noted between TT and CC genotypes (Table 4).
Table (4):
Comparative between number and percentage of follicles and oocytes that surrounded by CCs with genotypes.
| Parameters | T(114)C | G(129)R | G(199)A | ||||
|---|---|---|---|---|---|---|---|
| TT | CC | GG | GA | GG | AA | ||
| 1 | Follicles number with size (4-8mm) |
31 (44.28%) |
39 (55.71%) |
32 (45.71%) | 38 (54.28%) | 37 (52.85%) |
33 (47.14%) |
| Chi square | 1.644 NS | 4.327 * | 4.398* | ||||
| 2 | Oocytes surrounded completely and partially by CCs | 41 (46.59%) |
47 (53.40%) |
40 (45.45%) |
48 (54.54%) |
49 (55.68%) |
39 (44.31%) |
| Chi square | 3.381 NS | 4.278 * | 4.564 * | ||||
* (P<0.05)
Association between genotypes and ovarian histology
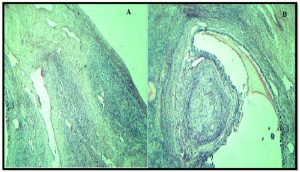
The ovarian tissues that obtained from sterile ewe’s specimens that possess A allele of G(199)A locus showed severe hypoplastic changes represented by diminished the number of developmental follicles (different phases of follicles are absent) that mostly replaced with stromal connective tissue (Fig.3A). In addition, the remnants follicles undergo severely degenerated follicular cells and proliferation of fibrous connective tissue which contained eosinophlic material (Fig. 3B).
Fig. 3. Ovarian histological sections for sterile ewes. (A) hypoplatic ovarian tissue, (B) follicular cells degeneration (H&E X40).
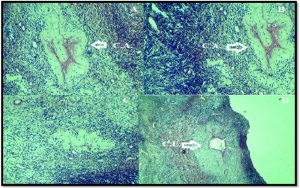
On the other hand, the ovarian histology that taken from fertile ewes that hada heterozygote G allele of G(145)R demonstrated normal follicular development with presence many corpus albicans in all samples. Moreover, presence degenerative corpus leutium (Fig. 4).
In this research, the polymorphic variations of coding GDF 9 gene were analyzed since that is an essential for folliculogenesis and female Awassi breed fertility.
By using sequence technique and analysis of the exon I of GDF-9 gene, three new variant SNPs were observed: T(114)C, G(129)R and G(199)A. Two of the three polymorphisms were nucleotide changes that do not result in an altered amino acid; Threonine (38) for T(114)C and glutamic acid (43) for G(129)R, The remaining nucleotide G(199)A gave rise to amino acid variation(conservative changes) negatively charged polar glutamic acid to positively charged polar lysine (67). These SNPs not be described before. By comparison the genotypes with phenotypic traits, the effect on fertility and infertility was noticeable.
These outcomes was in agreement with Bahrami et al. (2014), who find polymorphisms in exon I region of GDF9 gene in Hisari sheep by using restriction enzyme HhaI. Also agree with Hanrahan et al. (2004)study, which find that one SNP a missense 260G>A in exon I of GDF9 gene in Belclare and Cambridge Sheep breed(rs410123449), these SNPs substitutes one basic charged polar group Arginine to another polar charge (Histidine) at amino acid residue (87).
Association between genotypes, fertility and sterility
The outcomes of recent study (Table 3) showed that C mutant allele for T(114)C locus were noticed in sterile ewes, while wild T allele were fertile. Also, the A mutant allele for G(199)A locus were showed in sterile samples, while wild G allele related with fertile ewes. These SNPs not descripted before and agree with Hafezian (2011) study, which mentioned that presence C to T transitional mutation in exon I of Sangsari sheep and this related with reproduction (fertility and sterility).
Additionally, according to G (129) R, the sterile ewes have wild homozygote G allele, while fertile ewes possessed mutant heterozygote G allele. The recent study was in agreement with other reports reports (Davis et al. 2006; Galloway et al. 2000; Hafezian et al. 2011), they reported that homozygous genotype were sterile or reduced fertility rate. Additionally, our study agree with Barzegari et al. (2010), who showed that heterozygous for GDF9 (G>A) genotypes resulted in higher ovulation rates in Ghezeland Moghani Sheep, but not find association between mutation and sterility.
Moreover, the present study in agreement with Hanrahan et al. (2004), who conclude that the GDF9 SNPs in exon I and exon II were associated with increment ovulation rate,and subsequently responsible for enhancement the litter size in heterozygous allele for Belclare and Cambridge breeds ewes, but sterility in homozygotes. These events are happened due to failure of normal development of ovarian follicles.
The three SNPs that find in exon I reflect high effect on GDF9 function and thereafter effect on folliculogenesis, these facts are consistent with the Khodabakhshzadeh et al. (2016) results, which concluded that the high grade of genetic diversity that observed in ovine GDF9 gene coding region which probably affects folliculogenesis and ewe’s fertility.
Association between genotypes and assessment of follicles and oocytes
The current study find and confirmed the direct relationship between the SNPs of GDF9 with number of normal follicles and also with number of effective oocytes that been retrieved from ovaries. The proportion of follicles that recorded in mutant GA genotype (Table 4) were higher significant than GG in G(129)R. These views seem to be consistent with Mullen et al. (2014) finding, which suggested that the missense mutation in GDF9 diminishes the biologic effect of GDF9 mature protein and reducing its capacity to function as a feedback inhibitor to FSH, that may led to enhance ovulation rate in Finnsheep breed.
In addition, Vittet al. (2000) mentioned that in rats, the GDF9 inhibit FSH-induced cAMP synthesis, so the SNPin GDF9 of sheep may be cause increment the sensitivity of ovarian follicles to FSH, and that exert a positive effect on the number of developing follicle and CCsexpansibility. Moreover, GDF-9 decline mRNA synthesis of LH receptors, also it has particular impact on CCs via inducingCCsexpansion and improves hyaluronan synthase 2 (HAS2) and cyclooxygenase 2 (COX-2) expression. These events lead to rise of ovulation rate in G(129)R SNP.
Simultaneously, the finding noticed that mutant AA genotype of G(199)A locus recorded a marked diminish in follicles average number, as well as in oocytes number that enclosed by CCs. These results are in accord with Nicol et al. (2009) study indicating that homozygosity in oocyte GDF9 gene has negative effect in Thoka sheep breed (sterility).
Histological evaluation of both sterile and fertile ewes ovarian tissue showed that clearly histological differences between two groups,linkage between genotypes, sterility and fertility were approved through noticed the histological variation between different genotypes. As explained in the results, the follicular degeneration and hypoplasia were prevalent. Also, different phases of follicles are absent in case of mutant homozygote A allele of G(199)A locus. These outcomes corroborate the finding of Nicol et al. (2009) study, who suggested that ovaries from homozygous ewes were characterized by minimal follicles numbers that which beyond the primordial stage, also the major follicles that passing primary stage were abnormal and undergo degeneration. In fertile ewes the ovarian tissue that had heterozygote G allele of G(129)R the normal follicles were present, normal numerous corpus albicans and presence degenerative corpus leutium, these approved that the ewes were cyclic and fertile.
The present study concluded that the three SNPs in Awassi ewes have different effect on follicles and oocyte number, subsequently on fertility and sterility.
The homozygote mutation of G(199)A related with decrease follicles and oocyte number and with sterility, while the heterozygote mutation G(129)R related with normal fertility in Awassi ewes.
Acknowledgements
The authors thank the staff of laboratory of Biotechnology Research Center-Al-Nahrian University/Baghdad. Also special thanks for employers of Al-shola abattoir/ Baghdad.
Conflict of Interest
The authors declares that there is no conflict of interest.
- Al Barzinji, Y.M.S., Othman, G.U., 2013. Genetic Polymorphism in FecB Gene in Iraqi Sheep Breeds Using RFLP-PCR Technique. IOSR J. Agri. Vet. Sci. 2, 46-48.
- Asadpour, R., Jafari-Joozani, R., Alijani, S., Mahmod, H., 2012. Detection of polymorphism in booroola gene (FecB) and its association with litter size in Zel sheep breed in Iran. Slovak J. Anim. Sci. 45(2), 63-66.
- Bahrami, Y., Bahrami, S., Mohammadi, H.R., Chekani-Azar, V., Mousavizadeh, S.A., 2014.The Polymorphism of GDF-9 Gene in Hisari Sheep.In Biological Forum.6, 46. Res. Trend.
- Barzegari, A., Atashpaz, S., Ghabili, K., Nemati, Z., Rustaei, M., Azarbaijani, R., 2010. Polymorphisms in GDF9 and BMP15 associated with fertility and ovulation rate in Moghani and Ghezel sheep in Iran. Reprod. Domst. Anim. 45, 666-669.
- Bodensteiner, K.J., Clay, C.M., Moeller, C.L., Sawyer, H.R., 1999. Molecular cloning of the ovine growth/differentiation factor-9 gene and expression of growth/differentiation factor-9 in ovine and bovine ovaries. Biol. Reprod. 60, 381-386.
- Davis, G.H., 2005. Major genes affecting ovulation rate in sheep. Genetics Selection Evol.37, 11.
- Davis, G.H., Balakrishnan, L., Ross, I.K., Wilson, T., Galloway, S.M., Lumsden, B.M., Hanrahan, J.P., Mullen, M., Mao, X.Z., Wang, G.L., Zhao, Z.S., 2006.Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim.Reprod. Sci. 92, 87-96.
- De Castro, F.C., Cruz, M.H.C., Leal, C.L.V., 2016. Role of growth differentiation factor 9 and bone morphogenetic protein 15 in ovarian function and their importance in mammalian female fertility—A review. Asian-Aust. J. Anim. Sci. 29, 1065.
- Dong, J., Albertini, D.F., Nishimori, K., Kumar, T.R., Lu, N. and Matzuk, M.M., 1996. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 383, 531.
- Elvin, J. A., Yan, C., Matzuk, M. M., 2000. Oocyte-expressed TGF-β superfamily members in female fertility. Mol. Cell. Endocrinol. 159, 1-5.
- Elvin, J.A., Clark, A.T., Wang, P., Wolfman, N.M., Matzuk, M.M., 1999.Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol. Endocrinol. 13, 1035-1048.
- Fabre, S., Pierre, A., Mulsant, P., Bodin, L., Di Pasquale, E., Persani, L., Monget, P., Monniaux, D., 2006. Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reprod. Biol. Endocrinol. 4, 20.
- Galloway, S.M., McNatty, K.P., Cambridge, L.M., Laitinen, M.P., Juengel, J.L., Jokiranta, T.S., McLaren, R.J., Luiro, K., Dodds, K.G., Montgomery, G.W., Beattie, A.E., 2000. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nature genetics 25, 279.
- Hafezian, S. H., 2011. Genetic polymorphism BMP15 and GDF9 genes in Sangsari sheep of Iran. Int. J. of Genetics Mol. Biol. 3, 31-34.
- Hanrahan, J.P., Gregan, S.M., Mulsant, P., Mullen, M., Davis, G.H., Powell, R. and Galloway, S.M. (2004). Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovisaries). Biol. Reprod. 70, 900-909.
- Juengel, J.L., Hudson, N.L., Heath, D.A., Smith, P., Reader, K.L., Lawrence, S.B., O’Connell, A.R., Laitinen, M.P., Cranfield, M., Groome, N.P., Ritvos, O., 2002. Growth differentiation factor 9 and bone morphogenetic protein 15 are essential for ovarian follicular development in sheep. Biol. Reprod. 67, 1777-1789.
- Khodabakhshzadeh, R., Mohammadabadi, M.R., Esmailizadeh, A.K., Shahrebabak, H.M., Bordbar, F., Namin, S.A., 2016.Identification of point mutations in exon 2 of GDF9 gene in Kermani sheep. Polish J. Vet. Sci.19, 281-289.
- Kumm, K.I., 2008. Profitable Swedish lamb production by economic scale. Small Rum. Res. 81, 63-69.
- Lafi, S.Q., Talafha, A.Q., Giadinis, N., Kalaitzakis, E., Pourliotis, K., Panousis, N., 2009. Factors affecting the reproductive performance of Awassi sheep flocks in north-east of Jordan: An epidemiological study. Tropic. Anim. Health Prod., 41, 1755-1764.
- McNatty, K.P., Juengel, J.L., Reader, K.L., Lun, S., Myllymaa, S., Lawrence, S.B., Western, A., Meerasahib, M.F., Mottershead, D.G., Groome, N.P., Ritvos, O., 2005. Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function in ruminants. Reprod. 129, 481-487.
- Mullen, M. P., Hanrahan, J. P., 2014. Direct evidence on the contribution of a missense mutation in GDF9 to variation in ovulation rate of Finnsheep. PloS one. 9, 95251.
- Nicol, L., Bishop, S.C., Pong-Wong, R., Bendixen, C., Holm, L.E.,Rhind, S.M., McNeilly, A.S., 2009. Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep. Reprod. 138, 921-933.
- Notter, D. R., 2008. Genetic Aspects of Reproduction in Sheep.Reprod. Domest. Anim. 43, 122-128.
- Sadighi, M., Bodensteiner, K.J., Beattie, A.E., Galloway, S.M., 2002.Genetic mapping of ovine growth differentiation factor 9 (GDF9) to sheep chromosome 5. Anim. Genetics 33, 244-245.
- SAS., 2012. Statistical Analysis System, User’s Guide. Statistical.Version 9.1th ed. SAS.Inst. Inc. Cary.N.C. USA.
- Souza, C.J.H., McNeilly, A.S., Benavides, M.V., Melo, E.O., Moraes, J.C.F., 2014. Mutation in the protease cleavage site of GDF9 increases ovulation rate and litter size in heterozygous ewes and causes infertility in homozygous ewes. Anim. genetics 45, 732-739.
- Vitt, U.A., Hayashi, M., Klein, C., Hsueh, A.J., 2000. Growth differentiation factor-9 stimulates proliferation but suppresses the follicle-stimulating hormone-induced differentiation of cultured granulosa cells from small antral and preovulatory rat follicles. Biol. Reprod. 62, 370-377.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.