ISSN: 0973-7510
E-ISSN: 2581-690X
We discover essential enzymes catalyzing critical metabolic reactions as potential drug targets, which may help to fight Listeria infections and their associated secondary infections extensively and effectively. A comparative metabolic pathway approach has been applied to identify and determine putative drug targets against Listeria monocytogenes. For this, enzymes unique to pathogenic pathways of L. monocytogenes EGD-e were determined using the KEGG database. They were further refined by selecting enzymes with sequences non-homologous to the host Homo sapiens and analysing their essentiality to the pathogen’s survival. We report 15 essential pathogen-host non-homologous proteins as putative drug targets that can be exploited for development of specific drug targets or vaccines against multidrug resistant strains of L. monocytogenes. Finally, four essential enzymes from the pathogen: UDP-N-acetylglucosamine 1-carboxyvinyltransferase, Acetate kinase, Phosphate acetyltransferase, and Aspartate kinase were reported as novel putative targets for vaccine and drug development against L. monocytogenes infections. Unravelling novel target proteins and their associated pathways by comparing metabolic pathway analysis between L. monocytogenes EGD-e and host H. sapiens, develops the novelty of the work towards broad spectrum putative drug targets. This research design yields putative drug target critical enzymes that turn out to be fatal to the pathogen without interacting with the host machinery.
Listeria monocytogenes EGD-e, Homo sapiens, Comparative Metabolic Pathway Analysis, Critical Enzymes, Novel Putative Drug Targets
Humans are victim of a large number of microbial pathogens through infection in the respiratory tract, gastrointestinal tract, or genital tract epithelium. These pathogens further migrate to secondary locations through lymph and blood to cause diseases in the liver, spleen, bones, and central nervous system (CNS).1 Listeria monocytogenes is a Gram-positive, rod-shaped, foodborne pathogenic bacterium that is associated with listeriosis, CNS infection, meningitis, sepsis, liver infection, spleen infection and premature birth/ abortions.2,3 It is also associated with immune response based diseases like Rheumatoid Arthritis (RA) and Inflammatory Bowel Disease (IBD).4-6 L. monocytogenes has been found proliferating in colon of a patient suffering from IBD, much higher than in a healthy individual. Resistant biofilms of Listeria have been reported on all forms of synthetic and biological surfaces, including gut epithelium.7,8 L. monocytogenes enters the human host through the ingestion of contaminated food and reaches the intestine. It crosses the intestinal epithelial barrier and enters the bloodstream, from where it causes secondary infection in several organs such as liver, spleen, bones, brain etc.2 In South Africa, 978 people were found positive for listeriosis infection which led to 674 hospitalizations and 183 deaths were recorded in 2018.9 Listeriosis outbreaks due to food borne infections have been recorded regularly in Turkey.10 1,763 cases of listeriosis from 27 states were recorded by European Food Safety Authority (EFSA) in 2013, leading to 191 deaths.11 The fatality rate of listeriosis is about 20-30% among various parts of the world in 2018-19.12-14 The current treatment of such Listeria infections depends on the use of antibiotics, which turn out to be ineffective and have risks of developing drug-resistant strains. At first, the most common treatment for Listeria infections was administration of antibiotics such as penicillin G and/or ampicillin along with aminoglycoside such as gentamicin or kanamycin. Subsequently, a combination of trimethoprim and a sulphonamide was also used as a therapy.3,15 Initially, the majority of the L.monocytogenes strains (isolated from various sources) were susceptible to antibiotics, active against the Gram positive bacteria. The first drug resistance was observed against the antibiotic tetracycline in 1988.16,17 A multidrug resistant strain of L. monocytogenes was also isolated in France in 1988.16,17 Consequently, several multidrug resistant strains of Listeria have been discovered from environment, food sources and human listeriosis gut/stool samples.17,18 L. monocytogenes strains isolated from several food items (including ready-to-eat food) were reported to be resistant to several antibiotics such as ampicillin, lindamycin, nalidixic acid, penicillin (100%) and oxacillin (94.1%).19 The indiscriminate use of effective antibiotics in humans and animals has resulted in an exposure to high concentrations of these antibiotics, leading to resistance development in pathogenic strains of Listeria through gene alterations and transposons.20,21 The infection of these multidrug resistant strains of L. monocytogenes through various food sources may pose a serious threat to public health.9 So, this research attempts to find enzymes catalyzing essential metabolic reactions as potential drug targets, which may help to fight L. monocytogenes infections and their associated secondary infections extensively and effectively.
The easy accessibility of human genome sequences, complete genome sequences of several human infecting pathogens, and availability of several computational tools for their in silico analysis helps us to identify biomarkers and potential drug targets to combat these pathogens. Among these, comparative metabolic pathway analysis interpretations involve the understanding of the organism’s physiology, intracellular procedures, and networks, which can find the specific target molecules responsible for their survival, vital functions, or virulence. This can further be used to develop specific and effective drugs against the pathogens. Metabolic pathways illustrate how the bio-molecular units interact with each other to carry out the functions required for the survival, reproduction, and other organism-specific activities.22 Understanding the phenotype and function of organisms requires a detailed analysis of the metabolic pathways involved along with the study of single genes, as the expression of all the protein metabolites is a result of the action of enzymes catalyzing their interconversion. Proteins are the functional biomolecular element of the cell which converts the genetic information into practical reality. They are involved in gene regulation, cell metabolism, transport of nutrients, signal transduction, and enzymatic catalysis. Hence, the pathogen-specific proteins/enzymes can be used as broad-spectrum potential drug targets against these pathogens.
In the present study, comparative metabolic pathway analysis was implied between L. monocytogenes EGD-e (Reference genome) and H. sapiens with the aim to select enzymes unique to L. monocytogenes EGD-e, which are non-homologous to host. These enzymes can work as putative targets for the development of drugs to eradicate these pathogen without interacting with the host machinery. These enzymes were further analysed for their essential role in survival of the pathogen. As targeting the critical enzymes turn out to be fatal to the pathogen and help developing broad-spectrum drugs. Different comparative genomics and transcriptomics studies and comparative proteomic analysis have been performed between different strains of L. monocytogenes or between different species of Listeria with diverse objectives, but unravelling novel target proteins and their associated pathways by comparing metabolic pathway analysis between L. monocytogenes EGD-e and host Homo sapiens, develops the novelty of the work towards broad spectrum putative drug targets.
Metabolic Pathway Analysis of Pathogen and Host
Metabolic pathways of L. monocytogenes EGD-e and H. sapiens were retrieved from Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/).23 A manual assessment was performed to classify common and unique metabolic pathways between host and pathogen. The metabolic pathways present in pathogen but missing in the host were categorized as unique pathways, whereas the pathways present in both the pathogen and host were categorized as common pathways. The enzymes from unique metabolic pathways, having a sanctioned Enzyme Commission number (EC number), were mapped, and their protein sequences were extracted from the National Centre for Biotechnology Information (NCBI) genome database.24
Identification of Pathogen-Host Non-homologous Proteins
BLASTP25 analysis (Protein-protein Basic Local Alignment Search Tool) was performed for protein sequences of enzymes from unique metabolic pathways against the H. sapiens proteome. The protein sequences with an e value cut-off of 5e-3 were considered as homologous to the pathogen and were excluded from the study. The protein sequences without a hit under this criterion were considered to have no significant homolog in H. sapiens and selected for further analysis.26
Identification of Essential Pathogen-Host Non-homologous Proteins
BLASTP analysis was performed between the obtained non-homologous protein sequences and prokaryotic essential genes from database of essential genes (DEG; http://www.essentialgene.org/) to yield essential pathogen-host non-homologous proteins 27. The criteria of essentiality are as follows: evalue < 1e-10, bit score ≥ 100 and percentage identity ≥ 30.26
Subcellular Localization of Proteins
The exact position of these proteins in the cell was predicted using CELLO version 2.5 (subCELlular LOcalization predictor version 2.5).28 CELLO applies the amino acid composition and di-peptide composition based on physico-chemical parameters of amino acids to predict the subcellular position of the proteins.28 The gram-positive bacterial proteins have the following localization sites: the cell membrane, the cytoplasm, the extracellular space and the cell wall.
Structural Classification of the Unique Identified Target Proteins
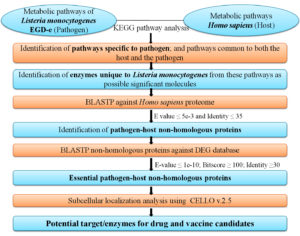
The predicted enzymes as drug targets were further analysed and classified for their three dimensional structure and presence of any best possible ligands in their binding pockets. The available structures of the predicted potential drug targets were retrieved and studied from the RCSB Protein Data Bank.29 The research design for this study has been presented in Figure.
Figure. Schematic representation of subtractive genomics methodology for identification of potential drug and vaccine targets against L. monocytogenes. KEGG pathway analysis of metabolism for the pathogen (L. monocytogenes) and the host (H. sapiens) led to the subsequent identification of essential pathogen-host non-homologous enzymes/proteins which can be targeted as possible drug/vaccine candidates. (KEGG: Kyoto Encyclopedia of Genes and Genomes; BLASTP: Protein-protein Basic Local Alignment Search Tool; DEG: Database of Essential Genes; CELLO v2.5: subCELlular LOcalization predictor version 2.5)
KEGG Pathway Analysis Interpretations
KEGG database analysis revealed a total of 107 pathways associated with L. monocytogenes EGD-e. Most of these pathways were biosynthesis and metabolic pathways such as glycolysis, TCA cycle, carbohydrate metabolism, fatty acid biosynthesis and degradation, amino acid biosynthesis and degradation, nucleotide metabolism, peptidoglycan biosynthesis, vitamin metabolism, central dogma, nitrogen and sulfur metabolism etc. Also, other pathways associated with bacterial functions and virulence such as secondary metabolites biosynthesis, microbial metabolism in different environments, degradation of aromatic compounds, resistance to antibiotics such as vancomycin and beta-Lactam, cationic antimicrobial peptide (CAMP) resistance, quorum sensing, chemotaxis, NOD-like receptor signaling pathway, two-component system, and bacterial invasion of epithelial cells were reported (https://www.genome.jp/kegg/).
H. sapiens have 337 reported pathways in the KEGG database. These pathways consist of the eukaryotic biosynthesis and metabolic pathways. They also have several pathways related to drug and xenobiotics resistance and metabolism, signalling pathways, apoptosis, muscle contraction, platelet activation, circadian cycle, hormone secretion and immune responses. A number of pathways were specifically related to diseases and infections such as Alzheimer disease, Parkinson disease, Huntington disease, prion diseases, drug addiction, Vibrio cholerae infection, Pathogenic Escherichia coli infection, Salmonella infection, malaria, tuberculosis, pathways in cancer, asthma, inflammatory bowel disease, and rheumatoid arthritis etc (https://www.genome.jp/kegg/).
The comparative metabolic pathway analysis using the KEGG database resulted in 28 pathways that were unique to the pathogen L. monocytogenes EGD-e, whereas 71 pathways, common to both L. monocytogenes EGD-e and H. sapiens (Table 1 and Table 2 respectively). A total number of 180 enzymes with valid EC numbers were identified from the unique pathogen pathways (Supplementary Table 1).
Table (1):
List of metabolic pathways common to both the pathogen Listeria monocytogenes EGD-e and host Homo sapiens (KEGG: Kyoto Encyclopedia of Genes and Genomes)
S. No. |
Metabolic pathway |
KEGG pathway ID |
|---|---|---|
1 |
Glycolysis / Gluconeogenesis |
lmo00010 |
2 |
Citrate cycle (TCA cycle) |
lmo00020 |
3 |
Pentose phosphate pathway |
lmo00030 |
4 |
Pentose and glucuronate interconversions |
lmo00040 |
5 |
Fructose and mannose metabolism |
lmo00051 |
6 |
Galactose metabolism |
lmo00052 |
7 |
Starch and sucrose metabolism |
lmo00500 |
8 |
Amino sugar and nucleotide sugar metabolism |
lmo00520 |
9 |
Pyruvate metabolism |
lmo00620 |
10 |
Glyoxylate and dicarboxylate metabolism |
lmo00630 |
11 |
Propanoate metabolism |
lmo00640 |
12 |
Butanoate metabolism |
lmo00650 |
13 |
Inositol phosphate metabolism |
lmo00562 |
14 |
Oxidative phosphorylation |
lmo00190 |
15 |
Nitrogen metabolism |
lmo00910 |
16 |
Sulfur metabolism |
lmo00920 |
17 |
Fatty acid biosynthesis |
lmo00061 |
18 |
Fatty acid degradation |
lmo00071 |
19 |
Synthesis and degradation of ketone bodies |
lmo00072 |
20 |
Glycerolipid metabolism |
lmo00561 |
21 |
Glycerophospholipid metabolism |
lmo00564 |
22 |
Arachidonic acid metabolism |
lmo00590 |
23 |
Purine metabolism |
lmo00230 |
24 |
Pyrimidine metabolism |
lmo00240 |
25 |
Alanine, aspartate and glutamate metabolism |
lmo00250 |
26 |
Glycine, serine and threonine metabolism |
lmo00260 |
27 |
Cysteine and methionine metabolism |
lmo00270 |
28 |
Valine, leucine and isoleucine degradation |
lmo00280 |
29 |
Valine, leucine and isoleucine biosynthesis |
lmo00290 |
30 |
Lysine degradation |
lmo00310 |
31 |
Arginine biosynthesis |
lmo00220 |
32 |
Arginine and proline metabolism |
lmo00330 |
33 |
Histidine metabolism |
lmo00340 |
34 |
Tyrosine metabolism |
lmo00350 |
35 |
Phenylalanine metabolism |
lmo00360 |
36 |
Tryptophan metabolism |
lmo00380 |
37 |
Phenylalanine, tyrosine and tryptophan biosynthesis |
lmo00400 |
38 |
beta-Alanine metabolism |
lmo00410 |
39 |
Taurine and hypotaurine metabolism |
lmo00430 |
40 |
Phosphonate and phosphinate metabolism |
lmo00440 |
41 |
Selenocompound metabolism |
lmo00450 |
42 |
D-Glutamine and D-glutamate metabolism |
lmo00471 |
43 |
D-Arginine and D-ornithine metabolism |
lmo00472 |
44 |
Glutathione metabolism |
lmo00480 |
45 |
Other glycan degradation |
lmo00511 |
46 |
Thiamine metabolism |
lmo00730 |
47 |
Riboflavin metabolism |
lmo00740 |
48 |
Vitamin B6 metabolism |
lmo00750 |
49 |
Nicotinate and nicotinamide metabolism |
lmo00760 |
50 |
Pantothenate and CoA biosynthesis |
lmo00770 |
51 |
Biotin metabolism |
lmo00780 |
52 |
Lipoic acid metabolism |
lmo00785 |
53 |
Folate biosynthesis |
lmo00790 |
54 |
One carbon pool by folate |
lmo00670 |
55 |
Porphyrin and chlorophyll metabolism |
lmo00860 |
56 |
Ubiquinone and other terpenoid-quinone biosynthesis |
lmo00130 |
57 |
Terpenoid backbone biosynthesis |
lmo00900 |
58 |
RNA polymerase |
lmo03020 |
59 |
Ribosome |
lmo03010 |
60 |
Aminoacyl-tRNA biosynthesis |
lmo00970 |
61 |
Protein export |
lmo03060 |
62 |
Sulfur relay system |
lmo04122 |
63 |
RNA degradation |
lmo03018 |
64 |
DNA replication |
lmo03030 |
65 |
Base excision repair |
lmo03410 |
66 |
Nucleotide excision repair |
lmo03420 |
67 |
Mismatch repair |
lmo03430 |
68 |
Homologous recombination |
lmo03440 |
69 |
ABC transporters |
lmo02010 |
70 |
NOD-like receptor signaling pathway |
lmo04621 |
71 |
Bacterial invasion of epithelial cells |
lmo05100 |
Table (2):
List of metabolic pathways which are unique to the pathogen Listeria monocytogenes EGD-e (KEGG: Kyoto Encyclopedia of Genes and Genomes)
S. No. |
Metabolic pathway |
KEGG pathway ID |
No. of proteins |
|---|---|---|---|
1 |
C5-Branched dibasic acid metabolism |
lmo00660 |
7 |
2 |
Methane metabolism |
lmo00680 |
17 |
3 |
Secondary bile acid biosynthesis |
lmo00121 |
2 |
4 |
Lysine biosynthesis |
lmo00300 |
17 |
5 |
Cyanoamino acid metabolism |
lmo00460 |
5 |
6 |
D-Alanine metabolism |
lmo00473 |
5 |
7 |
Peptidoglycan biosynthesis |
lmo00550 |
16 |
8 |
Polyketide sugar unit biosynthesis |
lmo00523 |
4 |
9 |
Carbapenem biosynthesis |
lmo00332 |
2 |
10 |
Monobactam biosynthesis |
lmo00261 |
6 |
11 |
Streptomycin biosynthesis |
lmo00521 |
7 |
12 |
Acarbose and validamycin biosynthesis |
lmo00525 |
2 |
13 |
Novobiocin biosynthesis |
lmo00401 |
3 |
14 |
Benzoate degradation |
lmo00362 |
3 |
15 |
Aminobenzoate degradation |
lmo00627 |
3 |
16 |
Chloroalkane and chloroalkene degradation |
lmo00625 |
1 |
17 |
Xylene degradation |
lmo00622 |
2 |
18 |
Styrene degradation |
lmo00643 |
3 |
19 |
Naphthalene degradation |
lmo00626 |
1 |
20 |
Phosphotransferase system (PTS) |
lmo02060 |
58 |
21 |
Bacterial secretion system |
lmo03070 |
3 |
22 |
Two-component system |
lmo02020 |
26 |
23 |
Quorum sensing |
lmo02024 |
12 |
24 |
Bacterial chemotaxis |
lmo02030 |
2 |
25 |
Flagellar assembly |
lmo02040 |
1 |
26 |
beta-Lactam resistance |
lmo01501 |
3 |
27 |
Vancomycin resistance |
lmo01502 |
6 |
28 |
Cationic antimicrobial peptide (CAMP) Resistance |
lmo01503 |
6 |
Determination of Essential Pathogen-Host Non-homologous Proteins
The unique pathogen enzymes were then compared with host proteome to identify host non-homologous proteins. Out of 180 enzymes, 120 enzyme sequences revealed d” 35% identity with human proteome, and 52 enzyme sequences did not show any hit against the human proteome (Supplementary Table 2). Enzymes which are unique to L. monocytogenes and also do not show any or significant homology to the host proteome may act as effective drug targets as these drug/vaccine candidates have reduced risk of any unwanted interaction with the host proteins. Hence, these drugs will be safe and not adversely affect the human host metabolism.
Essential genes are the least number of genes which are obligatory for the existence of any organism.30 Essential genes determined for 48 bacterial species have been listed in DEG. This essential genes list can be directly extracted and BLAST analysis can be performed. Alternatively, a common list of prokaryotic essential genes can be used for analysis of other organisms which are not listed in DEG, as performed in this study (DEG; http://www.essentialgene.org/). The knockout of any bacterial essential gene can produce lethal phenotypes, so essential genes may act as significant drug targets.31 This can also be exploited for development of specific drug targets or vaccines against multidrug resistant strains such as L. monocytogenes. Some essential genes may be conserved over a number of related species and are potential targets for development of broad spectrum antibiotics.27,31 Hence, 172 host-non-homologous enzyme sequences were analyzed for essentiality of L. monocytogenes using DEG. 98 enzymes were found to be essential enzymes of L. monocytogenes with an average identity of 49% to essential protein sequences of prokaryotes (Supplementary Table 3).
Subcellular Localization of Target Enzymes/Proteins
Subcellular localization of these enzymes was determined by CELLO v.2.5, which may provide important information about the function of protein. The bacterial proteins/enzymes present in Gram-positive bacterial dataset are mostly localized in the cytoplasm and the cell membrane. Rest of the proteins are localized in the extracellular space and very few are found at the cell wall.32 CELLO categorized our essential host non-homologous enzymes of L. monocytogenes, as presented in Supplementary Table 4.
Prediction of Potential Targets/ Enzymes for Drug and Vaccine Development
Out of the 52 non-homologous enzymes (with no identity match with H. sapiens proteome), 15 were recorded as essential enzymes for L. monocytogenes by DEG analysis with an average E value ≤ 1.4 e-23 and identity ≥ 47%. Further, we propose four L. monocytogenes enzymes as putative drug targets, completely non-homologous to human and critically important to survival of pathogen (with identity ≥ 60% to the essential prokaryotic sequences), as listed in Table 3. The nature and site of action of all these four enzymes was determined to be cytoplasmic. These enzymes tend to play a central role in the pathogenesis of L. monocytogenes, causing infections in humans.
Table (3):
Potential drug target enzymes from Listeria monocytogenesEGD-e, showing their essential role in survival of the pathogen and non-homologous nature to host Homo sapiens. KEGG: Kyoto Encyclopedia of Genes and Genomes)
S.No. |
Listeria monocytogenes EGD-e enzyme name |
KEGG ID |
Non homologous (Yes/No) and Percentage Identity (%) to Homo sapiens |
Essential (Yes/No) and Percentage Identity (%) to Listeria monocytogenesEGD-e |
Sub-cellular localization |
|---|---|---|---|---|---|
1 |
K00790 UDP-N-acetylglucosamine 1-carboxyvinyltransferase [EC:2.5.1.7] | |
lmo:lmo2526 |
Yes
0% |
Yes
72.55% |
Cytoplasmic |
2 |
K00925 acetate kinase [EC:2.7.2.1] | |
lmo:lmo1581 |
Yes
0% |
Yes
71.61% |
Cytoplasmic |
3 |
K00625 phosphate acetyltransferase [EC:2.3.1.8] | |
lmo:lmo2103 |
Yes
0% |
Yes
63.11% |
Cytoplasmic |
4 |
K00928 aspartate kinase [EC:2.7.2.4] | |
lmo:lmo1235 |
Yes
0% |
Yes
61% |
Cytoplasmic |
5 |
K02760 PTS system, cellobiose-specific IIB component [EC:2.7.1.196 2.7.1.205] | |
lmo:lmo2373 |
Yes
0% |
Yes
57.73% |
Cytoplasmic |
6 |
K02769 PTS system, fructose-specific IIB component [EC:2.7.1.202] | |
lmo:lmo0427 |
Yes
0% |
Yes
57.29% |
Cytoplasmic |
7 |
K04041 fructose-1,6-bisphosphatase III [EC:3.1.3.11] | |
lmo:lmo0830 |
Yes
0% |
Yes
51.85% |
Cytoplasmic |
8 |
K02799 PTS system, mannitol-specific IIB component [EC:2.7.1.197] |
lmo:lmo2799 |
Yes
0% |
Yes
50.11% |
Membrane |
9 |
K02777 PTS system, sugar-specific IIA component [EC:2.7.1.-] | |
lmo:lmo1017 |
Yes
0% |
Yes
48.41% |
Cytoplasmic |
10 |
K00887 undecaprenol kinase [EC:2.7.1.66] | |
lmo:lmo1464 |
Yes
0% |
Yes
46.4% |
Membrane |
11 |
K02755 PTS system, beta-glucoside-specific IIA component [EC:2.7.1.-] |
lmo:lmo2772 |
Yes
0% |
Yes
42.11% |
Membrane |
12 |
K00425 cytochrome bd ubiquinol oxidase subunit I [EC:7.1.1.7]| |
lmo:lmo2718 |
Yes
0% |
Yes
41.45% |
Membrane |
13 |
K01624 fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | |
lmo:lmo0359 |
Yes
0% |
Yes
39.1% |
Cytoplasmic |
14 |
K00003 homoserine dehydrogenase [EC:1.1.1.3] | |
lmo:lmo2547 |
Yes
0% |
Yes
38.26% |
Cytoplasmic |
15 |
K12555 penicillin-binding protein 2A [EC:2.4.1.129 3.4.16.4] | |
lmo:lmo2229 |
Yes
0% |
Yes
33.22% |
Membrane |
UDP-N-acetylglucosamine 1-carboxyvinyltransferase is an enzyme of class transferases that catalyzes the transfer of enolpyruvate group to UDP-N-acetyl-a-D-glucosamine which is a significant and committing reaction of cell wall formation in bacteria.33 It belongs to 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase family and subfamily of MurA. This enzyme also substantially involved in other biological processes like cell division, cell wall cycle, cell wall organization, and cell shape regulation along with peptidoglycan biosynthetic pathway.33,34 (https://www.uniprot.org/).
Acetate kinase is involved in the acetyl-CoA biosynthesis pathway by catalyzing a sub-pathway reaction of phosphorylating acetate utilizing ATP and a divalent cation such as Mg2+ or Mn2+. The reaction is summarized as: acetate + ATP = acetyl phosphate + ADP. The reverse reaction can also be catalyzed by this same enzyme. It is also involved in some metabolic intermediate biosynthesis, such as the organic acid metabolic process (https://www.genome.jp/kegg/). It has also been reported that acetyl phosphate levels in L. monocytogenes are directly involved in monitoring cell motility, chemotaxis, and resistant biofilm formation. In a study, Gueriri and co-workers developed acetate kinase mutants of L. monocytogenes with a blocked synthesis of acetyl phosphate. These mutants were reported with a phenotype of decreased ability of biofilm formation and diminished expression of flagellar protein biosynthesis and motility genes.35 Also, other studies have reported that some L. monocytogenes virulence factors such as VirR/VirS can be activated by the production of acetyl phosphate in the cells.36 The activity of acetate kinase has also been recorded in other pathogenic intestinal bacterial strains such as Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. These pathogen bacteria have been found to be involved in causing IBD in the human host.37
The third drug target enzyme, phosphate acetyltransferase, shows transferase activity. Phosphate acetyltransferase, along with the subsequent action of acetate kinase, produces acetate from acetyl-CoA (or acetyl phosphate) and generates ATP.38
The Aspartate kinase enzyme performs kinase activity, transferase activity, and binding of ATP by the reaction: ATP + L-aspartate = 4-phospho-L-aspartate + ADP. Aspartate kinase is involved in cellular amino acid biosynthetic pathways such as lysine biosynthesis via diaminopimelate (DAP) formation, homoserine biosynthesis, and threonine biosynthetic pathway 39 (https://www.uniprot.org/). These targets have not been used for drug/vaccine development to our best knowledge.
Other than these four novel drug targets, most of the other 11 enzymes have also been established as potential candidates for drug targets and have been reported in several other studies. Fructose-bisphosphate aldolase (FBA) class II, is a cytoplasmic or surface exposed bacterial enzyme catalysing the cleavage of fructose-1,6-bisphosphate to D-glyceraldehyde-3-phosphate and dihydroxyacetone phosphate, an important reversible step in glycolysis and gluconeogenesis.40 FBA is known to perform two or more unrelated functions in several bacterial species and hence is a moonlighting protein. FBA can play a significant role in binding to the host’s cells and to host’s proteins, subsequent generation of an immune response etc, and hence is involved in physiology and pathogenesis of the bacteria.40 The structure and sequence of FBA remains conserved among same and different bacterial species, so it can be exploited to develop broad spectrum antibiotics/vaccines against a wide group of pathogenic bacteria.41 Mendonca and coworkers reported FBA class II as a novel immunogenic surface protein and monoclonal antibody (mAb-3F8) against this protein for detection of the Listeria spp. and to distinguish Listeria from other pathogenic bacteria.42
The primary treatment for L. monocytogenes infected population is administration of b-lactam antibiotics (such as penicillins) which target a set of enzymes, the penicillin-binding proteins (PBPs) involved in peptidoglycan linking.43 Our findings also report penicillin-binding protein 2A as a putative drug target. Peptidoglycan is a foremost element of the bacterial cell wall which is integral to the cell structure and morphology. Peptidoglycan biosynthesis involves the creation of mesh like structure, which is facilitated by two steps: transglycosylation (elongation of glycan chain) and transpeptidation (peptide cross-linking the flanking glycan chains).44 High molecular weight Class A PBP catalyze both of these reactions through their N-terminal glycosyltransferase and C-terminal transpeptidase domain.44 Another research which applied several in silico approaches such as subtractive genomics and protein–protein interaction network topology, reported PDB4 along with 10 other proteins as a putative drug target in L. monocytogenes EGD-e proteome.45
We have also reported several phosphotransferase system (PTS) such as cellobiose-specific IIB component, mannitol-specific IIB component, fructose-specific IIB component, sugar-specific IIA component, and beta-glucoside-specific IIA component as putative drug targets. The PTS system is composed of a few soluble proteins and one membrane spanning protein, which are involved in the uptake/transport of PTS carbohydrates by the cell.46 It has also been reported to be actively involved in several regulatory functions such as catabolite repression, potassium transport, nitrogen and phosphate metabolism, antibiotic resistance, endotoxin production, biofilm formation, and virulence of several pathogens including L. monocytogenes.46 PTS mediated sugar transport of cellobiose, mannose, and glucose has been reported to regulate PrfA activity, which is in turn the major transcription factor regulating the virulence gene in L. monocytogenes.47 In a study based on comparative genomics of Vibrio cholera, several constituents of the PTS were described as drug and vaccine targets against the pathogen.48 Similarly, six components of the PTS system were reported as putative drug targets in Klebsiella pneumoniae MGH78578 using the in silico approach.49 Hence, the PTS system may act as an effective drug target for L. monocytogenes too.
Structural Classification of the Unique Identified Target Proteins
The available structures of the predicted potential drug targets were retrieved and studied from the RCSB Protein Data Bank.29 The structures of the predicted drug targets were available on PDB either in L. monocytogenes or in other microbes. The minimum criteria for considering the three dimensional structure was on the basis of Identity >= 70%, Query coverage of the sequence as 80% and E Value <= 0.00. The crystal structure of PBP 4 from L. monocytogenes in the Ampicillin bound form (PDB ID: 3ZG8)50 and PBP D2 from L. monocytogenes in apo form(PDB ID: 5ZQA)51 were accessible in PDB. Both these structures were studied in expression system of Escherichia coli. Several ligands such as (2r,4s)-2-[(1r)-1-{[(2r)-2-Amino-2-Phenylacetyl]amino}-2-Oxoethyl]-5,5-Dimethyl-1,3-Thiazolidine-4-Carboxylic acid, glycerol and Di(hydroxyethyl)ether have been reported to bind to the A and B chains of this enzyme. Structures for other drug target proteins were available for different other organisms. For instance, Apo structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. ‘Ames Ancestor’ (PDB ID: 3Q94) can be studied at PDB.52 This crystal structure was determined through X-ray diffraction experiment and the enzyme was found to be composed of A and B chains with two reported ligands (1,3-Dihydroxyacetonephosphate and acetate ion) interacting with the A chain of the protein. Similarly, the structure of Cytochrome BD-I ubiquinol oxidase from Escherichia coli (PDB ID: 6RX4) 53 and Homoserine Dehydrogenase from Saccharomyces cerevisiae (PDB ID: 1EBU) 54 were available. Two ligands (3-Aminomethyl-Pyridinium-Adenine-Dinucleotide and L-Homoserine) interacting with the D chain of the Homoserine Dehydrogenase enzyme and four ligands (Heme b/c, Cis-Heme d hydroxychlorin gamma-Spirolactone, 1,2-Dioleoyl-Sn-Glycero-3-Phosphoethanolamine, and Ubiquinone-8) binding with A and B chains of Cytochrome BD-I ubiquinol oxidase have been reported so far. Also, the structures of several enzymes reported as drug targets from the PTS system were retrieved for different organisms from PDB. Such as crystal structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis (PDB ID: 4MGE),55 and structure of IIB domain of the mannitol-specific permease enzyme II from Escherichia coli (PDB ID: 1VKR)56 can be studied from PDB. The crystal structure of the fructose specific IIB subunit of PTS system was available for Bacillus subtilis (PDB ID: 2R48).57 Similarly, the closest structures available for sugar-specific IIA component and beta-glucoside-specific IIA component of the PTS system were studied from PDB. The details of all these structures including the classification, expression system, mutations, gene names and ligand interactions have been compiled in Supplementary Table 5.
Out of the four novel drug targets predicted, the structures of two enzymes: UDP-N-acetylglucosamine 1-carboxyvinyltransferase and phosphate acetyltransferase were available for L. monocytogenes.58,59 The crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from L. monocytogenes EGD-e (PDB ID: 3R38) was determined through X-ray diffraction experiments in the Escherichia coli BL21 expression system. Two ligands have been determined for this protein (Sulfate ion and Chloride ion) which bind to the chain A of the protein.58 Similarly, the crystal structure of phosphate acetyl/butaryl transferase (from L. monocytogenes EGD-e) in complex with CoA (PDB ID: 3U9E) has been determined in Escherichia coli BL21(DE3) by X-ray diffraction studies. Four ligand molecules (Coenzyme A, Arginine, Glycerol, Chloride) have been known to interact with A and B chains of the protein59 (Supplementary Table 5).
The structure for the third novel protein acetate kinase was found for the organism Salmonella enterica subsp. enterica serovar Typhimurium (PDB ID: 3SK3) in the RCSB Protein Data Bank. Citric acid and 1,2-Ethanediol ligands interact with the A and B chain of the enzyme.60 Similarly, the structure for aspartate kinase was found for the organism Pseudomonas aeruginosa PAO1 (PDB ID: 5YEI) which was determined by X-ray diffraction in expression system Escherichia coli BL21(DE3). Three ligand molecules: Threonine, Lysine, and Glycerol are known to interact with the protein61 (Supplementary Table 5). Unpinning the structure categorization and identifying the inhibitors for these target proteins opened different methods of research towards drug design and reverse vaccinology approach.
The comparative metabolic pathway analysis approach of L. monocytogenes–H. sapiens resulted in four novel putative target proteins: UDP-N-acetylglucosamine 1-carboxyvinyltransferase, acetate kinase, phosphate acetyltransferase, and aspartate kinase, which were very high in essentiality index with the pathogen and non-homologous nature with the host. Other 11 enzymes on the list are also significant putative drug targets and some of them have been reported by prior studies as well. The predicted potential drug target enzymes from L. monocytogenes will not interact with host machinery and also perform essential functions such as peptidoglycan biosynthesis, cell motility, chemotaxis, resistant biofilm formation, virulence, bacterial pathogenicity, amino acid biosynthesis, cell division, and cell wall organization. Therefore, drug development against these targets to combat L. monocytogenes infections will be very promising. Unravelling novel target proteins and their associated pathways by comparing metabolic pathway analysis between Listeria monocytogenes EGD-e and host Homo sapiens, develops the novelty of the work towards specific and broad spectrum putative drug targets. In addition to this, a detailed further analysis of these potent target proteins in terms of in-vivo and in-vitro approach will attain new and unique generation of biomolecules against the diseases caused by L. monocytogenes.
Additional file: Additional Table S1- S5.
ACKNOWLEDGMENTS
The authors would like to thank Faculty of Biotechnology, Shri Ramswaroop Memorial University, Lucknow-Deva Road, Barabanki, India, for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NC and SS designed the experiments. NC performed the experiments. NC analyzed the data and wrote the manuscript. NC and TQ revised the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Clements A, Young JC, Constantinou N, Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes. 2012;3(2):71-87.
Crossref - Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018;16(1):32-46.
Crossref - Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55(3):476-511.
Crossref - Miranda-Bautista J, Padilla-Suarez C, Bouza E, Munoz P, Menchen L, Marin-Jimenez I. Listeria monocytogenes infection in inflammatory bowel disease patients: case series and review of the literature. Eur J Gastroenterol Hepatol. 2014;26(11):1247-1252.
Crossref - Diaz-Dilernia F, Costantini J, Nicolino TI, Sanchez MDL, Carbo L. Unusual Listeria monocytogenes hematogenous infection in total knee replacement treated with one-stage revision surgery. Arthroplast Today. 2019;5(3):296-300.
Crossref - Arulmozhi S, Matchado MS, Snijesh VP, Kumar A, Singh S. An insight into anti-arthritic property OF C25H34O7 for Rheumatoid arthritis using molecular modelling and molecular dynamics approach. Informatics in Medicine Unlocked. 2019;16:100145.
Crossref - Chiba M, Fukushima T, Koganei K, Nakamura N, Masamune O. Listeria monocytogenes in the colon in a case of fulminant ulcerative colitis. Scand J Gastroenterol. 1998;33(7):778-782.
Crossref - Chen W, Li D, Paulus B, Wilson I, Chadwick VS. Detection of Listeria monocytogenes by polymerase chain reaction in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. J Gastroenterol Hepatol. 2000;15(10):1145-1150.
Crossref - Sanlibaba P, Tezel BU, Cakmak GA. Prevalence and Antibiotic Resistance of Listeria monocytogenes Isolated from Ready-to-Eat Foods in Turkey. J Food Qual. 2018;7693782
Crossref - Erol Z, Tasci F. Overview of Listeria Monocytogenes as a Foodborne Pathogen: Traditional Review. Turkiye Klinikleri J Vet Sci. 2021;12(1):37-48.
Crossref - Ricci A, Allende A, Bolton D, et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA Journal. 2018;16(1):e05134.
Crossref - Zhang Y, Dong S, Chen H, et al. Prevalence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes From Retail Foods in Bulk in Zhejiang Province, China. Front Microbiol. 2019;10:1710.
Crossref - Li W, Bai L, Fu P, Han H, Liu J, Guo Y. The Epidemiology of Listeria monocytogenes in China. Foodborne Pathog Dis. 2018;15(8):459-466.
Crossref - Salama PJ, Embarek PKB, Bagaria J, Fall IS. Learning from listeria: safer food for all. Lancet. 2018;391(10137):2305-2306.
Crossref - Lorber B. Listeriosis. Clin Infect Dis. 1997;24(1):1-11.
Crossref - Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu AL, Courvalin P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet. 1990;335(8703):1422-1426.
Crossref - Charpentier E, Gerbaud G, Jacquet C, Rocourt J, Courvalin P. Incidence of antibiotic resistance in Listeria species. J Infect Dis. 1995;172(1):277-281.
Crossref - Abuin CMF, Fernandez EJQ, Sampayo CF, Otero JLI, Rodriguez LD, Saez AC. Susceptibilities of Listeria species isolated from food to nine antimicrobial agents. Antimicrob Agents Chemother. 1994;38(7):1655-1657.
Crossref - Arslan S, Ozdemir F. Prevalence and antimicrobial resistance of Listeria species and molecular characterization of Listeria monocytogenes isolated from retail ready-to-eat foods. FEMS Microbiol Lett. 2020;367(4):fnaa006.
Crossref - Iwu CD, Okoh AI. Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples. PLOS ONE. 2020;15(2):e0228956.
Crossref - Komora N, Bruschi C, Magalhדes R, Ferreira V, Teixeira P. Survival of Listeria monocytogenes with different antibiotic resistance patterns to food-associated stresses. Int J Food Microbiol. 2017;245:79-87.
Crossref - Oehm S, Gilbert D, Tauch A, Stoye J, Goesmann A. Comparative Pathway Analyzer—a web server for comparative analysis, clustering and visualization of metabolic networks in multiple organisms. Nucleic Acids Res. 2008;36(Suppl 2)):W433-437.
Crossref - Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42-46.
Crossref - Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35 suppl 1:D61-65.
Crossref - Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-3402.
Crossref - Sharma A, Pan A. Identification of potential drug targets in Yersinia pestis using metabolic pathway analysis: MurE ligase as a case study. Eur J Med Chem. 2012;57:185-195.
Crossref - Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37 suppl 1:D455-458.
Crossref - Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402-1406.
Crossref - Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-242.
Crossref - Koonin EV. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99-116.
Crossref - Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42 issue D1:D574-580.
Crossref - Wang X, Zhang J, Li GZ. Multi-location gram-positive and gram-negative bacterial protein subcellular localization using gene ontology and multi-label classifier ensemble. BMC Bioinformatics. 2015;16(Suppl 12):S1.
Crossref - van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18(5):503-519.
Crossref - Machata S, Hain T, Rohde M, Chakraborty T. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol. 2005;187(24):8385-8394.
Crossref - Gueriri I, Bay S, Dubrac S, Cyncynatus C, Msadek T. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol Microbiol. 2008;70(6):1342-1357.
Crossref - Mandin P, Fsihi H, Dussurget O, et al. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol. 2005;57(5):1367-1380.
Crossref - Kushkevych IV. Acetate kinase Activity and Kinetic Properties of the Enzyme in Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9 Intestinal Bacterial Strains. Open Microbiol J. 2014;8:138-143.
Crossref - Kelly AF, Patchett RA. Lactate and acetate production in Listeria innocua. Lett Appl Microbiol. 1996;23(2):125-128.
Crossref - Chen NY, Jiang SQ, Klein DA, Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268(13):9448-9465.
Crossref - Shams F, Oldfield NJ, Wooldridge KG, Turner DPJ. Fructose-1,6-bisphosphate aldolase (FBA)-a conserved glycolytic enzyme with virulence functions in bacteria: “ill met by moonlight.” Biochem Soc Trans. 2014;42(6):1792-1795.
Crossref - Elhaik Goldman S, Dotan S, Talias A, et al. Streptococcus pneumoniae fructose-1,6-bisphosphate aldolase, a protein vaccine candidate, elicits Th1/Th2/Th17-type cytokine responses in mice. Int J Mol Med. 2016;37(4):1127-1138.
Crossref - Mendonca M, Moreira GMSG, Conceicao FR, et al. Fructose 1,6-Bisphosphate Aldolase, a Novel Immunogenic Surface Protein on Listeria Species. PLoS ONE. 2016;11(8):e0160544.
Crossref - Nguyen UT, Harvey H, Hogan AJ, Afonso ACF, Wright GD, Burrows LL. Role of PBPD1 in stimulation of Listeria monocytogenes biofilm formation by subminimal inhibitory b-lactam concentrations. Antimicrob Agents Chemother. 2014;58(11):6508-6517.
Crossref - Haenni M, Majcherczyk PA, Barblan JL, Moreillon P. Mutational analysis of class A and class B penicillin-binding proteins in Streptococcus gordonii. Antimicrob Agents Chemother. 2006;50(12):4062-4069.
Crossref - Sarangi AN, Lohani M, Aggarwal R. Proteome mining for drug target identification in Listeria monocytogenes strain EGD-e and structure-based virtual screening of a candidate drug target penicillin binding protein 4. J Microbiol Methods. 2015;111:9-18.
Crossref - Deutscher J, Ake FMD, Derkaoui M, et al. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78(2):231-256.
Crossref - Mujahid S, Orsi RH, Boor KJ, Wiedmann M. Protein level identification of the Listeria monocytogenes sigma H, sigma L, and sigma C regulons. BMC Microbiol. 2013;13:156.
Crossref - Chawley P, Samal HB, Prava J, Suar M, Mahapatra RK. Comparative genomics study for identification of drug and vaccine targets in Vibrio cholerae: MurA ligase as a case study. Genomics. 2014;103(1):83-93.
Crossref - Georrge JJ, Umrania VV. In silico identification of putative drug targets in Klebsiella pneumonia MGH78578. Indian Journal of Biotechnology. 2011;10: 432-439
- Jeong JH, Kim YS, Rojviriya C, Ha SC, Kang BS, Kim YG. Crystal structures of bifunctional penicillin-binding protein 4 from Listeria monocytogenes. Antimicrob Agents Chemother. 2013;57(8):3507-3512.
Crossref - Jeong JH, Cha HJ, Kim YG. Crystal Structures of Penicillin-Binding Protein D2 from Listeria monocytogenes and Structural Basis for Antibiotic Specificity. Antimicrob Agents Chemother. 2018;62(9).
Crossref - Bank RPD. RCSB PDB – 3Q94: The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. “Ames Ancestor.” https://www.rcsb.org/structure/3Q94. Accessed August 1, 2020.
- TheBeling A, Rasmussen T, Burschel S, et al. Homologous bd oxidases share the same architecture but differ in mechanism. Nat Commun. 2019;10(1):5138.
Crossref - DeLaBarre B, Thompson PR, Wright GD, Berghuis AM. Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases. Nat Struct Biol. 2000;7(3):238-244.
Crossref - Bank RPD. RCSB PDB – 4MGE: 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis. Accessed August 1, 2020. https://www.rcsb.org/structure/4MGE
- Legler PM, Cai M, Peterkofsky A, Clore GM. Three-dimensional solution structure of the cytoplasmic B domain of the mannitol transporter IImannitol of the Escherichia coli phosphotransferase system. J Biol Chem. 2004;279(37):39115-39121.
Crossref - Bank RPD. RCSB PDB – 2R48: Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168. Accessed August 1, 2020.
Crossref - Bank RPD. RCSB PDB – 3R38: 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e. Accessed July 20, 2020. https://www.rcsb.org/structure/3R38
- Bank RPD. RCSB PDB – 3U9E: The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. Accessed July 20, 2020. https://www.rcsb.org/structure/3u9e
- Chittori S, Savithri HS, Murthy MRN. Structural and mechanistic investigations on Salmonella typhimurium acetate kinase (AckA): identification of a putative ligand binding pocket at the dimeric interface. BMC Struct Biol. 2012;12:24.
Crossref - Li CC, Yang MJ, Liu L, et al. Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase. Biochem J. 2018;475(6):1107-1119.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.