ISSN: 0973-7510
E-ISSN: 2581-690X
Coronavirus disease 2019 (COVID-19) is a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 has spread quickly across the world and has been declared a pandemic. Indonesia has many COVID-19 cases, with a high mortality rate. This study aimed to describe the distribution of COVID-19 in Indonesia and constructed the SARS-CoV-2 phylogenetic tree from Indonesian isolates and those from other countries, including other CoVs to determine their relationship. The distribution data of COVID-19 in Indonesia were obtained from the COVID-19 Management Handling Unit and descriptively analyzed. SARS-CoV-2 isolates were retrieved from the GenBank® (National Center of Biotechnology Information, USA) and GISAID EpiCoV™ databases and were used to construct phylogenetic trees using MEGA X software. Of the 37 provinces in Indonesia, five provinces with the highest case fatality rates were DKI Jakarta, Jawa Barat, Jawa Timur, and Banten, and the five provinces with the highest cure rate were Kepulauan Riau, Bali, Aceh, Gorontalo, and DI Yogyakarta. SARS-CoV-2 Indonesian isolates were closely related to SARS-CoV-2 isolates from other countries. The rapid and widespread distribution of SARS-CoV-2 in Indonesia was caused by the lack of compliance with territorial restrictions and dishonesty with medical personnel. These data revealed that mutations can occur during the transmission process, which can be caused by a history of travel and increased patient immunity.
COVID-19, Indonesia, Phylogenetic Tree, SARS-CoV-2
An unknown cause of acute respiratory disease occurred in Wuhan, China on December 2019 (Wong et al., 2020). The World Health Organization (WHO) established it as an outbreak and a global public health emergency on January 2020. On February 11, 2020, WHO named the disease COVID-19, caused by SARS-CoV-2, which was previously named as 2019-nCoV (Ghinai et al., 2020; Lai et al., 2020; World Health Organization, 2020a). WHO characterized COVID-19 as a pandemic on March 2020 (World Health Organization, 2020b), and as of May 9, 2020, COVID-19 has been informed in 215 countries with 3,822,382 confirmed cases and 263,658 mortalities (World Health Organization, 2020c), showing that the transmission of SARS-CoV-2 is very rapid.
SARS-CoV-2 is transmitted through droplets or direct contact (Lai et al., 2020). SARS-CoV-2 transmission route between people is direct transmission, such as sneezing, contact transmission, droplet inhalation, coughing, including contact with the mucous membranes of the nose, eyes, and mouth (Peng et al., 2020). In the process of transmission, SARS-CoV-2 infects humans due to the recognition of receptors; specifically, angiotensin-converting enzyme (ACE2) (Gheblawi et al., 2020; Shang et al., 2020; Zhang et al., 2020). SARS-CoV-2 enters cells through the bond between the spike glycoprotein and the host receptor, namely, ACE2 (Hoffmann et al., 2020; Zhang et al., 2020). ACE2 is present in various human organs and tissues (Hamming et al., 2004; Donoghue et al., 2000; Li et al., 2020)s evere acute respiratory syndrome coronavirus 2 (SARS-CoV-2; in the lungs, ACE2 is present in type II pneumocytes (Chen et al., 2010; Uhal et al., 2013).
The location of ACE2 in type 2 alveolar cells is the main cause of acute pneumonia in patients. The clinical symptoms of these patients include dry cough, fever, difficulty in breathing (dyspnea), an increase in body temperature, pneumonia, headache, a decrease in the number of white blood cells and lymphocytes, new pulmonary infiltrates visible upon chest radiography, and no noticeable improvement after medication with antibiotics for three days (Zhou et al., 2020) a large number of SARS-related coronaviruses (SARSr-CoVs. The worst outcome of SARS-CoV-2 infection is death.
On March 2, 2020, Indonesia reported the first case of SARS-CoV-2 infection in two Depok residents who had recently interacted with some Japanese citizens (Nuraini, 2020). Furthermore, COVID-19 began to emerge and was identified in various provinces in the territory of Indonesia, resulting in a high mortality rate in the country. As of May 9, 2020, Indonesia has a total of 13,645 confirmed COVID-19 cases, with 959 deaths and 2,607 patients declared cured (Worldometer, 2020; Gugus Tugas Percepatan Penanganan COVID-19, 2020). The wide distribution of COVID-19 allows mutations to occur in the transmission process. The genetic diversity of the virus little by little increases as the virus spreads; certain genome variants have been repeatedly sequenced at different geographical locations and time points (Skums et al., 2020). Phylogenetic trees are usually built to determine the evolutionary relationships between species (Horiike, 2016) it is important to choose the suitable molecule for the phylogenetic analysis of a given lineage. For example, when the evolutionary rate of the gene (or protein. This study aimed to describe the distribution of COVID-19 in Indonesia and also constructed the phylogenetic tree of Indonesian SARS-CoV-2 isolates with isolates from other countries and other CoVs to determine their relationship.
Distribution of COVID-19 in Indonesia
The COVID-19 case data in Indonesia were collected from the Gugus Tugas Percepatan Penanganan COVID-19 database until May 8, 2020. The data were analyzed descriptively to determine the five provinces with the highest case fatality rate (CFR) and cure rate levels.
SARS-CoV-2 Isolates
The sequences of SARS-CoV-2 isolates were retrieved from the GenBank® (National Center of Biotechnology Information (NCBI), USA) and GISAID EpiCoV™ databases (Table 1).
Table (1):
SARS-CoV-2 and other CoV isolates from the NCBI and GISAID databases.
Code |
Origin |
Host |
Isolation Source |
|---|---|---|---|
MT188341.1 |
USA: Minnesota (MN) |
Homo sapiens |
Nasopharyngeal or oropharyngeal swab |
MN908947.3 |
China: Wuhan |
Homo sapiens |
Unknown |
MN985325.1 |
USA: Washington (WA) |
Homo sapiens |
Oropharyngeal swab |
MN988713.1 |
USA: Illinois (IL) |
Homo sapiens |
Sputum |
MT253701.1 |
China: Zhejiang, Hangzhou |
Homo sapiens |
Sputum |
MN994468.1 |
USA: California (CA) |
Homo sapiens |
Nasopharyngeal swab |
EPI_ISL_414377 |
Indonesia: Jakarta |
Homo sapiens |
Sputum |
EPI_ISL_414376 |
Indonesia: Jakarta |
Homo sapiens |
Sputum |
EPI_ISL_414375 |
Indonesia: Jakarta |
Homo sapiens |
Sputum |
EPI_ISL_413219 |
Indonesia: Jakarta |
Homo sapiens |
Oro-pharyngeal swab |
Molecular Phylogenetic Analysis
The phylogenetic model and tree visualization were constructed based on the SARS-CoV-2 nucleocapsid protein gene using MEGA X with the maximum likelihood method. The phylogenetic tree was validated by running the analysis on 1000 bootstrapped input datasets using the Tamura-Nei substitution model.
Distribution of COVID-19 in Indonesia
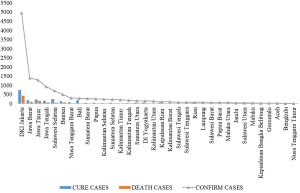
Fig. 1 shows the distribution of COVID-19 cases in 34 provinces in Indonesia. From the surveillance system report conducted by the acceleration of COVID-19 countermeasures, the confirmed cases of COVID-19 have spread to all provinces. The five provinces with the highest distribution of confirmed cases were DKI Jakarta, Jawa Barat, Jawa Timur, Jawa Tengah, and Sulawesi Selatan. These five provinces also had a relatively high number of deaths. The number of cases cured in the five provinces was also considerably high. The province of Nusa Tenggara Timur (NTT) reported the least number of confirmed cases (n=12). Additionally, no deaths have been reported in NTT. Other provinces that have not reported deaths are Maluku Utara and Jambi.
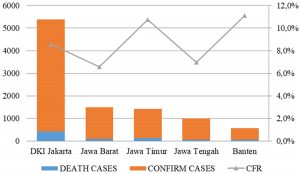
On May 8, 2020, 13,122 confirmed cases were reported in Indonesia. The cases consisted of 13,091 confirmed cases from 34 provinces and an additional 21 cases that were being verified in the field. The number of reported deaths from 31 provinces was 943 (CFR=7.2%) (Gugus Tugas COVID-19, 2020), which was obtained from a COVID-19 surveillance report conducted in Indonesia. If the surveillance system attributes are examined, namely the completeness of the data, the data only reaches approximately 30% for reporting positive confirmed (35%) and cured cases (39%), and 67.7% for deaths. All confirmed cases (positive, death, or recovered) should have had a reporting date, but many officials did not include a complete report date.
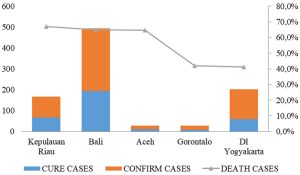
Indonesia has a high mortality rate due to COVID-19. The five provinces in Indonesia with the highest CFR were Banten (11.1%), Jawa Timur (10.7%), DKI Jakarta (8.6%), Jawa Tengah (7%), and Jawa Barat (6.6%). The calculation of mortality was obtained by comparing the number of deaths with the number of confirmed cases (Fig. 2). In addition, DKI Jakarta was the largest contributor to COVID-19 cases in Indonesia and was also the point of entry of SARS-CoV-2 in Indonesia. This was evident from Indonesia’s index case originating from DKI Jakarta. DKI Jakarta has the largest international airport in Indonesia, which is the entrance for foreign tourists. A positive index case occurred after contact with Japanese citizens visiting Indonesia. From DKI Jakarta, the virus spread to the surrounding provinces, especially Jawa Barat. A cluster from Jakarta spread to other areas in the vicinity due to the large number of residents from Jawa Barat who work in Jakarta (DetikNews, 2020). The main coping effort for COVID-19 was the selection of cases. Cases that were identified in the community received treatment. Treatments that have been administered in Indonesia are shown in Fig. 3. Kepulauan Riau, Bali, Aceh, Gorontalo, and DI Yogyakarta were the five provinces that reported the highest cure rates.
The probability of transmission of SARS-CoV-2 is that one positive person can transmit the virus to two to three other people, with an average transmission time of five days (DetikNews, 2020). According to epidemiology theory (Gordis, 2004), this is referred to as the R0 (R-naught). COVID-19 cases in Indonesia occurred by spreading from metropolitan cities, with the main source of transmission being the Jabodetabek cluster, and then to Surabaya, which is the second largest metropolitan city in Indonesia, as well as the capital of Jawa Timur Province (Erfinanto, 2020). Metropolitan cities are considered sources of employment and education. The largest cluster of SARS-CoV-2 transmission in Jawa Timur was the Hajj Cluster in Sukolilo, Surabaya (Kurniawan, 2020). This is the main cause of the acceleration of the spread of cases in Indonesia (KompasTV, 2020; Mantra, 2000).
The mortality rate (CFR) in Indonesia is relatively high. This can be observed from Indonesia’s CFR (CFR=7.2%), which exceeds that of the world (6.9%) (Kementrian Kesehatan, 2020). The diverse character of Indonesian society was the main reason for the difficulty in preventing and controlling the spread of SARS-CoV-2. Not all Indonesian people adhered to local lockdowns or territorial restrictions, which are the main mechanisms for suppressing the spread of SARS-CoV-2. Additionally, there were many people who were sick and not honest when checking themselves into health facilities. This is precisely the source of SARS-CoV-2 transmission to medical personnel (Liputan 6, 2020).
Phylogenetic Tree Construction
SARS-CoV-2 is thought to have originated from bats that migrated to humans through food in a wholesale market for seafood and wild animals near the city of Wuhan, China (Hui et al., 2020; Li et al., 2020). Bats of the genus Rhinolophus in southern China are known to have many SARS-like CoVs (Sun et al., 2020a). SARS-CoV-2 belongs to the subgenus Sarbecovirus, genus Betacoronavirus, and family Coronaviridae (Gorbalenya et al., 2020). SARS-CoV-2 has characteristics similar to SARS-CoV, including its genome sequence (Xu et al., 2020). SARS-CoV-2 is also similar to the Middle East respiratory syndrome-related coronavirus (MERS-CoV) (Elfiky et al., 2017). This group of viruses is genetically diverse and shows frequent recombination (Sun et al., 2020b; Zhou et al., 2020).
In the distribution process, SARS-CoV-2 may mutate, as SARS-CoV-2 is an RNA virus. The characteristic of RNA viruses is that they mutate up to 1 million times faster than their host (Pachetti et al., 2020). Phylogenetic analysis is a method to explain the evolutionary history and relationships among a group of organisms (Horiike, 2016) and is usually visualized in the form of phylogenetic trees to make it easier to determine the relationship between species. In this study, phylogenetic trees were constructed using the Indonesian SARS-CoV-2 nucleocapsid gene sequences with sequences from isolates sourced from several other countries, as well as sequences of several other types of CoVs collected from the NCBI and GISAID databases. The N-protein is the most abundant protein in coronavirus (Fig. 1), is encoded by a conserved gene, and is used as a molecular marker (Sino Biological, 2020).
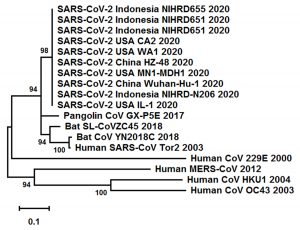
Fig. 4. Phylogenetic tree of SARS-CoV-2 and other CoV isolates based on the nucleocapsid protein gene
Fig. 4 shows that SARS-CoV Tor2 2003 has the closest relationship with Bat-CoV YN2018C 2018. SARS-CoV-2 2020, which is found in various countries, is also closely related to Pangolin-CoV GX-P5E 2017 and Bat-SL-CoV ZC45 2018. The bat coronavirus (BatCoV RaTG13), previously detected in Rhinolophus affinis from Yunnan province, has many similarities with SARS-CoV-2, with an overall genome sequence similarity of 96.2% (Zhou et al., 2020). SARS-CoV-2 2020 is also related to MERS-CoV 2012, Human-CoV HKU1 2004, and Human-CoV OC43 2003; SARS-CoV-2 has a genetic similarity of 50% with MERS-CoV and 79% with SARS-CoV (Rabaan et al., 2020). SARS-CoV and SARS-CoV-2 genome sequences have extremely high levels of nucleotide similarity with only six genomic regions showing differences (Xu et al., 2020).
SARS-CoV-2 from in Indonesia was very closely related to SARS-CoV-2 from other countries. This demonstrated that they had very similar genomes, although there were slight genetic changes during the transmission process. The results of the study by Forster et al. (2020) show that the patient’s travel history determines patterns in the SARS-CoV-2 phylogenetic network. Different phylogeographic patterns can be influenced by different migration histories, founder events, and sample sizes. Additionally, patient treatment and increased patient immunity also affects mutation occurrence. Any decrease in COVID-19 cases and/or CFR is likely caused by increased immunity in the human population and is an epidemiological context of viral mutation (Zhang and Holmes, 2020). Monitoring mammalian viruses is very important to prevent the occurrence of virus transmission from animals to humans. Furthermore, monitoring viruses that have infected humans is important to prevent wider transmission and to identify mutations during transmission.
Of the 37 provinces in Indonesia, five provinces with the highest CFR were DKI Jakarta, Jawa Barat, Jawa Timur, and Banten, and five provinces with the highest cure rate were Kepulauan Riau, Bali, Aceh, Gorontalo, and DI Yogyakarta. The rapid and widespread distribution of SARS-CoV-2 in Indonesia was caused by the lack of compliance with territorial restrictions and dishonesty with medical personnel. SARS-CoV-2 isolates from Indonesia were closely related to SARS-CoV-2 isolates from other countries. These data revealed that mutations can occur during the transmission process, which can be caused by a history of travel and increased patient immunity.
ACKNOWLEDGMENTS
This study supported by the Directorate General of Higher Education, Ministry of Education and Culture of the Republic of Indonesia.
CONFLICT OF INTEREST
The authors declares that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study funded by the Directorate General of Higher Education, Ministry of Education and Culture of the Republic of Indonesia.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Chen IY, Chang SC, Wu HY, et al. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J Virol. 2010;84(15):7703-7712.
Crossref - DetikNews. Kapan Sebenarnya Corona Pertama Kali Masuk RI. 2020.
- Donoghue M, Hsieh F, Baronas E, et al. A novel angiotensin-converting enzyme-related carboxy-peptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87(5):E1-E9.
Crossref - Elfiky AA, Mahdy SM, Elshemey WM. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J Med Virol. 2017;89(6):1040-1047.
Crossref - Erfinanto E. Update Situasi Corona COVID-19 di Jawa Timur pada 1 April 2020 – Surabaya Liputan6.com. 2020.
- Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. PNAS. 2020;117(17):9241-9243.
Crossref - Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ Res. 2020;126(10):1456-1474.
Crossref - Ghinai I, McPherson TD, Hunter JC, et al. Illinois COVID-19 Investigation Team. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395(10230):1137-1144.
Crossref - Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536-544.
Crossref - Gordis L. Epidemiology. Philadelphia: Elsevier Inc; 2004.
- Gugus Tugas COVID-19. Peta Sebaran | Gugus Tugas Percepatan Penanganan COVID-19; 2020.
- Gugus Tugas Percepatan Penanganan COVID-19. Data Sebaran. Retrieved May 9, 2020, from https://covid19.go.id
- Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631-637.
Crossref - Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.e8.
Crossref - Horiike T. An introduction to molecular phylogenetic analysis. Rev Agri Sci. 2016;4:36-45.
Crossref - Hui DS, Azhar EI, Madani TA, et al. The continuing epidemic threat of novel coronaviruses to global health – the latest novel coronavirus outbreak in Wuhang, China. Int J Infect Dis. 2020;91:264-266.
Crossref - Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2):244.
Crossref - Kementrian Kesehatan. Pedoman COVID REV-4. Pedoman Pencegahan Dan Pengendalian Coronavirus Disease (COVID-19), 1 (Revisi ke-4), 2020; 1-125.
- KompasTV. Penambahan Kasus COVID-19 Tembus 533, Jawa Timur Paling Banyak; 2020.
- Kurniawan D. Penjelasan Pemprov Jatim Terkait Klaster Baru di Bojonegoro – Surabaya Liputan6; 2020.
- Lai CC, Liu YH, Wang CY, Wang YH, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J Microbiol Immunol Infect. 2020;S1684-1182(20)30040-2.
Crossref - Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924.
Crossref - Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9(1):45.
Crossref - Li X, Wang W, Zhao X, et al. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92(5):501-511.
Crossref - Liputan6. 5 Kisah Kebohongan Pasien Sebabkan Tenaga Medis Terinfeksi Corona – News Liputan6; 2020.
- Mantra IB. Demografi Umum. Yogyakarta: Pustaka Pelajar; 2000.
- Nuraini R. Kasus COVID-19 Pertama, Masyarakat Jangan Panik | Indonesia.go.id. Retrieved May 6, 2020, from https://indonesia.go.id/narasi/indonesia-dalam-angka/ekonomi/kasus-COVID-19-pertama-masyarakat-jangan-panik; 2020
- Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, Masciovecchio C, Angeletti S, Ciccozzi M, Gallo RC, Zella D. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179.
Crossref - Peng X, Xu X, Li Y, Cheng L, Zhou X, Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int J Oral Sci. 2020;12(1):9.
Crossref - Rabaan AA, Al-Ahmed SH, Haque S, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez Med. 2020;28(2):174-184.
- Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020.
Crossref - Sino Biological. SARS-Cov-2 (2019-nCoV) Antigen Reagents. Retrieved May 7, 2020, from https://www.sinobiological.com/research/virus/2019-ncov-antigen; 2020
- Skums P, Kirpich A, Baykal PI, Zelikovsky A. Global transmission network of SARS-CoV-2 : from outbreak to pandemic. medRxiv. 2020;2020.03.22.20041145.
Crossref - Sun J, He WT, Wang L, et al. COVID-19: Epidemiology, Evolution, and Cross-Disciplinary Perspectives. Trends Mol Med. 2020;26(5):483-495.
Crossref - Uhal BD, Dang M, Dang V, et al. Cell cycle dependence of ACE-2 explains downregulation in idiopathic pulmonary fibrosis. Eur Respir J. 2013;42(1):198-210.
Crossref - Wong MC, Cregeen SJJ, Ajami NJ, Petrosino JF. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020;2020.02.07.939207.
Crossref - World Health Organization. Coronavirus (COVID-19) events as they happen. Retrieved May 6, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen; 2020a.
- World Health Organization. Coronavirus disease (COVID-19) Pandemic. Retrieved March 28, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019; 2020b.
- World Health Organization. Coronavirus disease (COVID-19) outbrake situation. Retrieved May 7, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019; 2020c.
- Worldometer. Coronavirus Cases. Worldometer. https://doi.org/10.1101/2020.01.23.20018549V2; 2020.
- Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586-590.
Crossref - Zhang YZ, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell, 2020;181(2):223-227.
Crossref - Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.