ISSN: 0973-7510
E-ISSN: 2581-690X
Insights into the molecular pathogenesis of respiratory viral infections were investigated using serum and peripheral blood from patients with clinical syndromes. Signatures of expression of cytokines, genes and apoptotic proteins that discriminate symptomatic individuals from healthy individuals were determined among 21 patients. In symptomatic patients, significant upregulation of IL-1β, IL-2, IL-4, IL-6, IL-8, IL-12, IL-15, TNF-a and IFN-g (P<0.05) was noted, while IL-10 was significantly downregulated (P<0.05). This is accompanied by either up or down-regulation of various pro-apoptotic and anti-apoptotic markers, suggesting a protective role of immune responses against viral infection and the capacity of viruses to subvert host cell apoptosis. Gene expression analysis for both T and B cells were categorized according to their functional status of activation, proliferation, and differentiation. Of note, genes SH2D1A and TCL1A were upregulated only in rhinovirus samples, while PSMB7, CD4, CD8A, HLA-DMA, HLA-DRA and CD69 were upregulated in samples of Flu A and RSV but were not significant in samples of rhinovirus as compared to healthy individuals. These results demonstrated Flu A and RSV elicit different alterations in human peripheral blood gene expression as compared to rhinovirus. Overall, despite the small number of study subjects, the current study for the first time has recognized signature genes, cytokines and proteins that are used by some respiratory viruses that may serve as candidates for rapid diagnosis as well as targets for therapeutic interventions.
PBMC, cytokines, respiratory viruses, gene expression
Respiratory viruses are major causes of morbidity and mortality in humans and represent an economic burden worldwide. Influenza A (Flu A), rhinovirus (RV) and respiratory syncytial virus (RSV) are identified as important public health problems.1Although different respiratory viruses represent distinct genome structure, exclusive entry receptors and replication approaches, they cause similar clinical syndromes such as fever, sneezing, sore throat, cough, rhinorrhea and coryza.
The respiratory tract is a vulnerable site for infection of respiratory viruses and has its own defence strategies in the elimination of pathogens. A complicated response initiated by resident respiratory tract cells and innate immune cells, and ultimately by adaptive immune cells, is involved in viral clearance.2 Pathogen associated molecular patterns (PAMPs) on the virus is recognized by the recognition receptors (PRRs) of the innate immune system and this includes toll-like receptors (TLRs), retinoic-acid-inducible gene-I (RIG-I)-like receptors (RLRs), and nucleotide binding-oligomerisation domain (NOD)-like receptors (NLRs). PRRs are expressed in pulmonary epithelial cells and the ligands activate epithelial cells in order to initiate a rapid immune response against viral invasion.3 Some viruses avoid recognition by TLRs but are recognizable by RIG-I4 or the cytosolic NLR NOD2.5 Many viruses possess common signalling pathways and hence this elicits conserved outcomes with triggering of Type I IFNs which is a central feature together with nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) mediated regulation of various inflammatory cytokines.6 This anti-viral state includes resistance to viral replication in all cells by induction of apoptotic cell death in infected cells, increased major histocompatibility complex (MHC) class I expression, activation of dendritic cells (DCs) and macrophages, and stimulation of natural killer (NK) cells to enhance their cytolytic activity.7 At the initial stage of the innate immune reaction, inflammatory cytokines such as IL-1β, IL-6, IL-12 and TNF-a are also produced. These cytokines increase endothelial expression of adhesion molecules, increase vascular permeability, and further activates the adaptive immune
response.8 Macrophages play a critical role in phagocytosis of infected cells and release chemokines, such as CXCL9 and CXCL10, which then results in recruitment of inflammatory cells. Dendritic cells loaded with viral peptides migrate to secondary lymphoid organs, interact with naïve CD4+ and CD8+ T lymphocytes, activating them resulting in differentiation of these T cells into effectors which then migrate into the circulation. These activated T cells also interact with B cells which then causes the production of various classes of antibody with specific neutralizing effects on viruses.6 Complement is also a critical component of anti-viral immunity. Altogether, these mechanisms of the adaptive immune response result in viral clearance.
An ultimately efficient response against viral infection is lysis of infected cells through apoptosis which play a protective role by blocking the virus from completing its replication. However, some viruses have evolved the capacity to avoid apoptosis. On the other hand, apoptosis can be beneficial for viruses as it helps to disseminate the virus progeny.9 The communication between innate and adaptive immunity, and also apoptosis activation involves important molecules such as cytokines and chemokines. Despite extensive research on the immune responses against respiratory viruses, the exact mechanisms underlying the induction of innate and adaptive immune responses against viruses is still unknown. Understanding the host immune responses to these viral infections enables us to not only understand disease pathobiology and helps towards diagnostics for these infections. Viral clearance needs a complex and multi-faceted immune response by the resident respiratory tract cells which is then finally eliminated by the adaptive immune system which completes viral clearance by eliminating infectious virions.10
Gene expression profiling obtained from peripheral blood mononuclear cells (PBMCs) enable distinguishment between the immune modifiers,11 the physiologic states12 exposures to pathogens13 and also to environmental exposures.14 Currently, viral infections are diagnosed by detection of pathogen.15 Development of pathogen-specific signatures from gene expression of whole blood and analyzing the differences by advanced statistical methods for analysis of data sets may enable it possible to classify the infections by profiling the genes expressed by the host. In this study, we developed peripheral blood gene expression signatures which were extracted from respiratory viral infected humans to evaluate the different genes involved in adaptive immunity against respiratory viral infection. In addition, levels of 14 cytokines and 43 apoptosis proteins were measured to get a better understanding of the interaction between the virus and the host immune response.
Ethics statement and patient samples
Blood samples from twenty-one patients (10 males and 11 females, with a median age of 28 years (range of 18-40)) with confirmed respiratory tract infection by FTD Respiratory Pathogens 21 Assay (FTD Resp 21) (Fast Track Diagnostics, Luxembourg) were used in this study. FTD Resp 21 allows the detection for the presence of adenovirus (HAdV), bocavirus (HboV), coronavirus 229E (HCoV-229E), coronavirus HKU1 (HCoV-HKU1), coronavirus NL63 (HCoV-NL63), coronavirus OC43 (HCoV-OC43), enterovirus (EV), metapneumovirus A/B (MPV), parechovirus (HPeV), influenza A (Flu A), influenza A virus subtype H1N1 (H1N1), influenza B (Flu B), Mycoplasma pneumoniae, parainfluenza virus 1 (PIV1), parainfluenza virus 2 (PIV2), parainfluenza virus 3 (PIV3), parainfluenza virus 4 (PIV4), respiratory syncytial virus A/B (RSV), rhinovirus (RV). The blood collected was separated for serum and peripheral blood mononuclear cell (PBMC). PBMC were isolated by Ficoll separation and cryopreserved in liquid nitrogen within 8 h of blood draw. All data analyzed were anonymized and ethical clearance for this work was approved by the Scientific and Ethical Committee of University of Malaya Medical Centre (UMMC) (Ethics Committee/ IRB Reference No: 20143-60).
Identification and Quantification of Cytokines
Fourteen different cytokines namely IL-1β, IL-2, IL-4, IL-6, IL-7, IL-8, IL-10, IL-12, IL-13, IL-15, IL-18, TNF-a, IFN-γ, and GS-CSF were simultaneously measured in the serum samples of patients using Cytokine/Chemokine Magnetic Bead Panel, MCYTOMAG-60K (Milliplex MAP Kit, Millipore Corporation, USA) according to the manufacturer’s instructions. Briefly, patients’ serum samples were incubated with beads coated with antibodies (Abs) to various cytokines, each having a unique fluorescent intensity. The mixtures were then incubated with biotinylated anti-cytokine antibodies, followed by PE-conjugated streptavidin. The fluorescent signals were measured using the MAGPIX multiplex reader. Raw data was determined as the relative fluorescence intensity and then converted to cytokines concentration based on the standard curve generated from the reference concentrations supplied in the kit.
Identification and Quantification of human apoptosis proteins
Forty-three human apoptosis protein levels in patient sera were evaluated using the Raybio® Human Apoptosis Antibody Array (Norcoss, GA, USA) kit according to the manufacturer’s instructions. The list of human apoptosis antibodies included in the array are namely BAD, BAX, Bcl-2, Bcl-w, BID, BIM, Caspase3, Caspase8, CD40, CD40L, clap-2, cytoC, DR6, Fas, FasL, HSP27, HSP60, HSP70, HTRA, IGF-I, IGF-II, IGFBP-1, IGFBP-II, IGFBP-3, IGFBP-4, IGFBP-5, IGFBP-6, P21, P27, P53, SMAC, Survivin, sTNF-R1, sTNF-R2, TNF-a, TNF-β, TRAILR-1, TRAILR-2, TRAILR-3, TRAILR-4, IGF-1sR, livin, and XIAP. Briefly, 100µl of each sample were added to the antibody array membranes and incubated for 2 h at RT. Samples were removed and each well was washed using Wash Buffer I and Wash Buffer II. Seventy microliters of diluted biotin-conjugated antibodies were added to each well followed by incubation at RT for 2 h. After washing, 70 µl of Hilyte PlusTM-conjugated streptavidin was added to each subarray. Finally, the slides were incubated for 2 h at RT in dark room and washed with Wash Buffer II. The signals were imaged using Axon Gene Pix 4000B (Molecular Devices, Sunnyvale, CA, USA). The data was analysed using the RayBio® Analysis Tool (RayBiotech, Norcross, USA).
Gene expression profiling by NanoString
Samples detected with respiratory viruses (influenza A, respiratory syncytial virus, and rhinovirus) were targeted for gene expression analysis of PBMC using NanoString gene expression profiling assay. For this purpose, RNA was extracted using a Qiagen RNeasy Plus mini kit following the manufacturer’s protocol. RNA yield and quality were evaluated on a NanoDrop 2000 (Thermo Scientific). The NanoString technology measures RNA abundance at the single molecule level and was performed using adaptive immunity panel of NanoString kit (NanoString, USA) according to the manufacturer’s instructions. RNA is the starting material for this assay and forms complexes with capture and reporter probes which are then captured on a solid surface and imaged. Molecule counts are then generated. The reporter consists of a string of seven fluorochromes, with four different colors available to fill each position. Each reaction contained 250 ng of total RNA in a 5 μL aliquot, reporter and capture probes, 6 pairs of positive control and 8 pairs of negative control probes. Analysis and normalization of the raw NanoString data was conducted using nSolver Analysis Software v1.1 (NanoString, USA). Raw counts were normalized to internal levels of 7 reference genes: CNOT2, GAPDH, HPRT1, PHGDH, SUMO2, SYS1, and WDR45L. A background count level was estimated using the average count of the 8 negative control probes in every reaction plus 2 standard deviations.
Statistical Analysis
ANOVA analysis was used to determine differentially expressed genes (significant P value < 0.05) using the Minitab software (Minitab, USA).
Serum cytokine profile
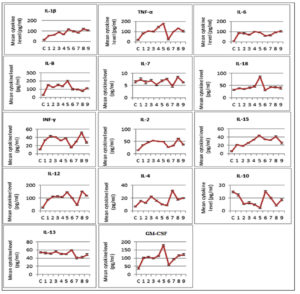
The levels of 14 different types of cytokines in patients with respiratory tract infections were determined. In general, from 21 viruses detected samples, 17 samples showed significant expression levels of the cytokines compared with the healthy individuals. Specifically, significant upregulation in the expression levels of IL-1β, IL-6, IL-8, IL-12, IL-2, IL-15, IL-4, TNF-α, IFN-γ and GS-CSF (P<0.05) was observed in the virus detected samples including Flu A, H1N1, Flu B + H1N1, RV + H1N1, RV + RSV + EV, RV, RSV and HAdV. Samples with Flu A, H1N1, Flu B + H1N1, RV + H1N1, RSV, HAdV and triple detection of RV, RSV, EV viruses showed a significant downregulation of IL-10 (P<0.05). The sample of Flu B + H1N1 showed the most significant results for IL-1β, IL-2, IL-8, IL-12, IL-15, IL-10, TNF-a, and GS-CSF. Additionally, IL-18 was significant only in groups with Flu B + H1N1. No significant results were observed in the expression level of IL-7 in all groups (Fig. 1).
Fig. 1. Mean level of 14 cytokine including IL-1β, TNF-a, IL-6, IL-8, IL-7, IL-18, INF-γ, IL-2, IL-15, IL-12, IL-4, IL-10, IL-13 and GS-CSF in virus detected serum samples compared with control (C). Samples 1 to 9: (1) Influenza A, (2) Influenza A (subtype H1N1), (3) Influenza A + Influenza A (subtype H1N1), (4) Rhinovirus + Influenza A (subtype H1N1), (5) Influenza B + Influenza A (subtype H1N1), (6) Rhinovirus, (7) Respiratory syncytial virus, (8) Rhinovirus + Respiratory syncytial virus + Enterovirus, (9) Adenovirus.
Apoptosis proteins expression profile
Based on the human apoptosis antibody array, 43 apoptosis proteins in 21 respiratory virus detected samples were analysed. Values above 2 and below 0.65-fold change were considered as up- and down- regulation, respectively. In samples with a single pathogen detection, pro-apoptotic and anti-apoptotic proteins were affected with highest frequency of bcl-2 antagonist of cell death (BAD), bcl-2 like protein 11(BIM), bcl-2 like protein 4(BAX), Cluster of differentiation 40 (CD40), CD 40Ligand (CD40L) , an initiator cysteine aspartic acid proteases (Caspase 3 and Caspase 8), p27, p53, TRAILR-1, TRAILR-2 in samples with Flu A, H1N1, RV and RSV. In samples with dual pathogens (RV + H1N1, Flu B + H1N1, PIV3 + PIV4) some of pro-apoptotic and anti-apoptotic proteins were upregulated with higher frequencies of BAD, BAX, CD40, CD40L, Caspase3, sTNF-R1, sTNF-R2, TRAILR-1, TRAILR-2, p21 and p53. In the one sample with 3 pathogens, RV + RSV + EV, upregulation of BAD, BIM, BAX, CD40, CD40L, cytoC, HSP27, IGFBP-4, TRAILR-1 and TRAILR-2 was observed (Table 1).
Table (1):
Up- and down-regulated apoptosis proteins in virus detected serum samples compare with healthy individuals.
| Virus Detected Samples | Up-Regulated Apoptosis Proteins | Down-Regulated Apoptosis Proteins |
|---|---|---|
| Single pathogen | ||
| Influenza A virus | BAD, BIM, BAX, CD40, CD40L, Caspase 3, Caspase 8, IGFBP-1, IGFBP-4, p27, p53, sTNF-R2, TRAILR-1, TRAILR-2 | bcl-w, bcl-2, HSP60, HSP70, livin, SMAC, Survivin, cytoC, HTRA, p21 |
| Influenza A virus (subtype H1N1) | BAD, BIM, CD40L, cytoC, FasL, Caspase 3, Caspase 8, HTRA, IGF-II, IGFBP-1, IGFBP-3, p53, sTNF-R2, TRAILR-1, TRAILR-2 | bcl-2, bcl-w, HSP27, IGFBP-2, IGF-1sR, livin, Survivin, CD40, p27 |
| Human rhinovirus | BAD, BIM, BAX, CD40L, cytoC, HTRA, Caspase 3, Caspase 8, IGFBP-1, IGFBP-3, p21, p27, p53, TRAILR-1, TRAILR-2 | bax, bcl-w, BIM, cytoC, HSP60, HSP70, IGFBP-5, IGF-1sR, livin, SMAC, Survivin, XIAP |
| Human respiratory syncytial virus A/B | BAD, BIM, BAX, CD40, CD40L, cytoC, FasL, Caspase 3, Caspase 8, HTRA, IGF-II, IGFBP-4, p21, p27, p53, sTNF-R1, sTNF-R2, TRAILR-1, TRAILR-2 | bcl-2, bcl-w, cIAP-2, HSP60, HSP70, IGF-II, IGF-1sR, livin, sTNF-R1, XIAP |
| Human adenovirus | BAD, BIM, BAX, CD40, CD40L, HSP27, p27, TRAILR-1, TRAILR-2 | bax, bcl-w, HSP27, HSP60, HSP70, IGF-I, IGF-II, IGF-1sR, Livin, Survivin, XIAP, IGFBP-3, IGFBP-4 |
| Dual Pathogens | ||
| Influenza A virus (subtype H1N1) Influenza B | BAD, BIM, BAX, Caspase3, CD40, CD40L, cytoC, FasL, HTRA, IGF-II, p21, p53, sTNF-R1, sTNF-R2, TRAILR-1, TRAILR-2 | bcl-w, CD40, cIAP-2, HSP27, HSP60, HSP70, IGF-I, IGF-II, IGF-1sR, Livin, Survivin, XIAP, IGFBP-4, p27 |
| Human parainfluenza virus 3 Human parainfluenza virus 4 |
BIM, Bax, Caspase 3, p21, p27, TRAILR-1, TRAILR-2 HSP27, sTNF-R2 | bcl-2, bcl-w, BIM, cIAP-2, IGF-1sR, livin, Survivin, CD40, CD40L, cytoC, HTRA |
| Human rhinovirus Influenza A virus (subtype H1N1) |
BAD, BAX, Caspase3, CD40, CD40L, cytoC, FasL, HTRA, IGF-II, p21, p53, sTNF-R2, TRAILR-1, TRAILR-2 | bcl-2, bcl-w, HSP60, HSP70, IGF-1sR, livin, Survivin, XIAP, IGFBP-1, p27 |
| Triple Pathogens | ||
| Human rhinovirus Human respiratory syncytial virus A/B Enterovirus |
BAD, BIM, BAX, CD40, CD40L, cytoC, HSP27, IGFBP-4, TRAILR-1, TRAILR-2 | bcl-2, bcl-w, HSP60, HSP70, IGF-I, IGF-II, IGF-1sR, livin, Survivin, sTNF-R1, XIAP |
| Human bocavirus Human coronavirus HKU1 Human coronavirus 229E |
NIL | bcl-2, bcl-w, cIAP-2, HSP27, HSP60, HSP70, IGF-II, IGF-1sR, livin, SMAC, Survivin, BIM, CD40L, cytoC |
However, it was also observed that some proteins such as B-cell lymphoma 2 (bcl-2), bcl-w, cellular inhibitor of apoptosis proteins 2 (cIAP-2), heat-shock proteins (HSP27, HSP60, HSP70), Insulin-like growth factors (IGF-II), livin, second mitochondrial activator of caspases (SMAC) and Survivin were downregulated in samples with single pathogen detected (Flu A, H1N1), RSV, RV and HAdV). Livin, bcl-2, bcl-w, HSP60, HSP70, IGF-1sR and Survivin were down-regulated in samples with RV + H1N1 and Flu B + H1N1. In the sample with PIV3 and PIV4 viruses, besides downregulation of bcl-2, bcl-w, BIM, IGF-1sR, livin and Survivin, reduced levels of CD40, CD40L, cytoC, HTRA was also observed. In samples with 3 pathogens (HboV + HCoV-HKU + HCoV-229E, and RV + RSV + EV), downregulation of bcl-2, bcl-w, HSP60, HSP70, Survivin, livin, IGF-II, IGF-1sR was detected (Table 1).
Evaluation of genes associated with B cells activation, proliferation and differentiation
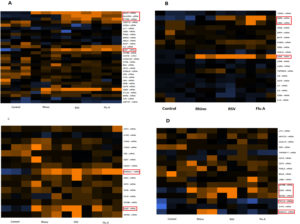
Fifteen percent of lymphocytes are B-lymphocytes that function primarily in humoral immunity in healthy adults.16 The criteria for B-cell activation, B cell function, B cell signalling, and B-cells maturation was targeted to evaluate ‘B-cell signature’ genes. As compared to healthy individuals’ genes including CD69, CD38, LRBA, CD27, PTP RC, HLA-A, HLA-DRA, CD79B, CD22, TCL1A and SH2D1A as B-cell signature genes were significantly upregulated in samples of Flu A, RSV or rhinoviruses. Specifically, B cell function and signalling genes such as CD27, PTP RC, HLA-A and HLA-DRA were significantly upregulated in samples of Flu A and RSV as compared to the control. Samples with rhinovirus showed upregulation of CD79B, TCL1A and SH2D1A as well as CD27, CD38, HLA-A. In the category of genes involved in activation of B cells, upregulation of genes CD69, CD38 and LRBA were observed in samples with Flu A and RSV viruses as compared to healthy individuals while rhinovirus samples showed significance in CD38. B cell maturation genes CD22 and stbGal were upregulated in all samples as compared to control (Fig. 2). The results also indicate that genes CD69 and LRBA were upregulated among samples of RSV and Flu A and not in samples of rhinovirus.
Fig. 2. Expression of B-cell signature genes for all samples (Flu A, RSV, RV, Control). A: Genes involved in B cell function; B: genes involved in B cell activation; C: genes involved in B cell maturation; D: Genes involved in B cell signalling. Expression levels are represented by a colour key in which bright yellow represents the highest levels and bright blue represents the lowest levels, and less saturated shades represent intermediate levels of expression. Red boxes indicate the upregulation is significant.
Evaluation of genes associated with T cells activation, proliferation, and differentiation
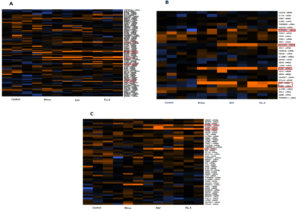
Seventy percent of lymphocytes are T-lymphocytes that function primarily in cell-mediated immunity (Palmer et al., 2006). T-cell signature genes were evaluated by seeking out genes that met the following involvement: i) Activated T cell, ii) T cell function and iii) T cell signalling. Totally, CD5, CD74, CXCR3, CXCR4, CD8B, PRS6, KLRB, LCP and AN32B, TNRS4, CD38, IL12RB1, PRF1 and HLA-DRA, NCK1, CD40LG, CD96, VAV1 and CCl4 were upregulated in samples of Flu A, RSV, and RV. Specifically, in category of T cell function genes significant upregulation in the expression levels of CD5, CXCR3, CXCR4, CD8B, PRS6 and CD2 were observed in the virus detected samples of Flu A and RSV. In this category, samples with RV showed significant upregulation of KLRB. Evaluation of genes involved in T cell activation indicated genes TNRS4 and CD38 were upregulated in samples of Flu A and RSV while genes HLA-DRA and IL12RB1 were upregulated only in rhinovirus samples (Fig. 3).
Fig. 3. Expression of T-cell signature genes for samples of Flu A, RSV, RV, Control. A: genes involved in T cell function; B: genes involved in activation of T cells; C: genes involved in T cell signalling. Expression levels are represented by a colour key in which bright yellow represents the highest levels and bright blue represents the lowest levels, and less saturated shades represent intermediate levels of expression. Red boxes indicate upregulation of genes.
Evaluation genes associated with antigen presentation
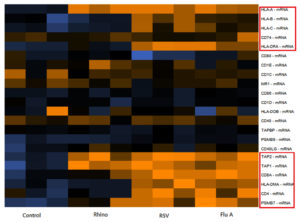
In mammalian cells antigen presentation. Followed by T cell activation are central processes in the cell-mediated immune responses. Different genes involved in antigen presentation including CD86, CD74, CD8A, CD40LG, CD1D, CD4, TAP1, PSMB7, TAPBP, HLA-DOB, CD40, TAP2, HLA-DMA, CD28, CD1C, HLA-A, PSMB9, HLA-B, HLA-C, HLA-DRA, and MR1 were evaluated in respiratory viral infected samples. Among them, CD8A, CD4, TAP 1, PSMB7, TAP2, HLA-DMA, HLA-A, HLA-B, HLA-C, HLA-DRA, CD74 showed significant upregulation in samples of Flu A and RSV as compared to control. In rhinovirus samples only the HLA-A, TAP1 and TAP2 genes were upregulated (Fig. 4).
Fig. 4. Expression of different genes involved in antigen presentation for samples of Flu A, RSV, RV and Control. Expression levels are represented by a colour key in which bright yellow represents the highest levels and bright blue represents the lowest levels, and less saturated shades represent intermediate levels of expression. Red boxes indicate upregulation of genes.
Pathway enrichment analysis
Bases on the gene expression profile, it was found that the CD4, CD8A, TAP1, TAP 2, HLA-A, HLA-B, HLA-C, HLA-DMA and HLA-DRA were differentially elevated among patients infected by flu and RSV; whilst VAV1, CCl4, KLRB1, KLRD1, LCP1, AN32B, FCER2, TCL1A and SH2D1A were differentially elevated among patients infected by rhinovirus. Pathway enrichment analysis of these two sets of genes revealed that Flu and RSV infection were associated with antigen processing/presentation pathway, while rhinovirus infection was associated with NK cell activation pathway.
This study on human respiratory viral infections including Flu A, RSV and RV was carried out to determine gene expression patterns of host-based peripheral blood specific to the adaptive immune responses to these viral infections. For this purpose, 192 genes related to adaptive immunity were evaluated in PBMCs extracted from respiratory viral infected patients. In order to simplify the characterization of analysis, genes were categorized into three groups including genes associated with activation, proliferation and differentiation of T cells, genes associated with activation, proliferation and differentiation of B cells, and genes associated with antigen presentation. Results provide evidence that gene expression of peripheral blood can potentially be considered as biomarkers for specific infectious pathogens and may function as efficient diagnostic tools for triaging treatment decisions for respiratory infection. Triage and treatment decisions can be facilitated by distinction between infectious causes of illness. Although traditional culture and PCR based diagnostics are effective in determining aetiology, these techniques have some limitations especially the high false negative rate when the test is performed during the window period or late when immune response had cleared the virus in circulation.17 Classification of infectious pathogens according to host immune responses has potential to increase our diagnostic abilities and to provide additional understanding into the infection pathobiology. Woods et al18 has shown that, in H1N1 and H1N3 infections, the peripheral blood gene expression signature can distinguish between these two strains with good degree of accuracy. Additionally, their study demonstrated that genomic signatures were able to distinguish upper respiratory viral infection from pneumonia due to Streptococcus pneumoniae.19
There are different types of T cells that are distinguishable based on their cell surface markers and functional capabilities. These T cells that play a regulatory role in adaptive immunity, whether cellular or humoral immune responses, are referred to as T helper (Th) cells and typically express CD4+ molecules on their surface. Activation of virus specific CD4+ Th cells, recognize virus-derived MHC class II-associated peptides on antigen presenting cells. Viral epitopes associated with MHC class I molecules activate the naïve CD8+ T cells in the draining lymph nodes, which subsequently differentiates into cytotoxic T lymphocytes (CTLs).20 Different gene sets encompass many areas of the T-cell receptor together with the associated signal transduction genes and this also include genes encoding transcriptional regulators and cell adhesion molecules.21,22
In this study, we found that some genes are common among different respiratory viral infections; whilst some genes were differentially elevated in different respiratory viral infections. We found that genes related to cell adhesion and costimulatory molecules including CD40, CD40L, CD2, CD86, CD38, CD28 were the common genes upregulated in respiratory viral infected. It has been shown that costimulatory signals enable amplification of the T-cell receptor (TCR)-induced signal with a limited number of MHC–peptide complexes. With CD28 costimulation, the number of TCR molecules decreases and these require to be engaged on a single T cell to stimulate activation and phosphorylation (Noble, 2000). Additionally, other genes that were upregulated include TNFRS4, IL12RB1, PRF1, CXCRS in all samples of Flu A, RSV and RV. These genes tightly regulate the threshold, magnitude and duration of T cell responses against respiratory viral infections.26,6
In another analysis, we found genes including CD4, CD8, TAP1/2, HLA-DMA, HLA-A/B/C, HLA-DRA were significantly increased in samples of Flu A and RSV. The protein encoded by TAP1/2 genes is associated with assembly of the class I molecules through transfer of cytosolic peptides to the endoplasmic reticulum into the membrane-bound compartment.24 HLAs including A, B and C, present peptides from inside the cell. HLA-DMA and HLA-DR (HLA class II alpha chain paralogues) is a heterodimer consisting of an alpha (DMA) and a beta chain (DMB), both anchored in the membrane and found in intracellular vesicles. The DM molecule helps to release the CLIP molecule from the peptide binding site. Genes critical in B cell activation, function and signalling such as CD69, CD38, LRBA and CD27 as were also upregulated in samples of RSV and Flu A.25,26
On the other hand, genes such as VAV1, CCL4, KLRB1, KLRD1, LCP1, AN32B, FCER2, TCL1A and SH2D1A were upregulated in samples of rhinovirus only. Among these genes, VAV1, KLRB1, KLRD1, AN32B and SH2D1A were found to be involved in NK cell activation. VAV1 was initially identified as a proto-oncogene,4 and later was showed to played a central role in NK cell-mediated cytotoxicity8. The both KLRB1 and KLRD1 are two classes of killer lectin-line receptor, while SH2D1A is an adaptor protein that involve in the activation of NK cell.5 While KLRB1 (also known as CD161) is an early differentiation marker for NK cells and act as an activation receptor;27 KLRB1 (also known as NKG2A) is an inhibitory receptor that interrupt the activation of NK cells,28,29 suggesting that perhaps this is the viral strategy to evade from NK cell surveillance. Intriguingly, FCER2 gene that encoded for Fc epsilon receptor 2 was also found to be upregulated among rhinovirus-infected patients. Given that Fcε is the receptor for IgE, this is at least in part explained why rhinovirus has often associated with allergic responses such as asthma among infected children.30,31,9 In general, gene expression analysis revealed significant alterations of different genes among Flu A, RSV, and RV samples respectively. Respiratory viral infection stimulates alterations in gene expression of human peripheral blood cells that SH2D1A and TCL1A genes were upregulated only in samples of rhinovirus. Additionally, different kinds of genes such as PSMB7, CD4, CD8A, HLA-DMA, HLA-DRA and CD69 were upregulated in samples of Flu A and RSV while they were not significant in sample of RV compared to healthy individuals.
When viruses infect the cells of the respiratory tract, the host reacts by activating parallel signalling pathways that then leads to transcription of cytokine genes essential for activation of the adaptive immune responses.31 Programmed cell death or apoptosis is another strategy of the cells to eliminate viral infection11. Viruses developed several strategies to evade the immune defence mechanism against infection. On the other hand, viruses profit from apoptosis as dissemination of virus progeny can occur via induction of the death of the infected cells.32 Therefore, our experiment was also designed to describe alterations in the expression level of some important cytokines and apoptosis proteins in patients with clinical syndromes of respiratory infection.
Significant increase in the levels of IL-1β, IL-2, IL-4, IL-6, IL-8, IL-12, IL-15, TNF-a, and IFN-γ in all resultant groups showed that infection of studied respiratory viruses resulted in a cascade of signalling events leading to the expression of pro-inflammatory, type 1 T helper (Th1) and type 2 T helper (Th2) cytokines which may be of importance for viral clearance. It has been investigated that the presence of many respiratory viruses including RSV29, RV14, and influenza27 with single stranded RNA cause to activate the TLR3 or TLR8. Moreover, double-stranded RNA and CpG DNA can be recognized by TLR7 and TLR9, respectively10. TLR7 and TLR9 use MyD88 as an essential adaptor for the activation of NF-B and IRF-7 which leads to the production of proinflammatory cytokines33 and type I IFNs15. The RIG-I and MDA5 (melanoma differentiation-associated gene 5) induces an antiviral response and this has been seen with RSV where it stimulates replication-mediated activation of the IFN-β promoter via RIG-I signalling22. Small RNA viruses such as rhinoviruses can be detected by MDA5 and it is very important in transcription of IFN- β, IFN- λ and proinflammatory cytokines26. Additionally, negative-sense ssRNA viruses, such as influenza virus and RSV are detectable by RIG-I and play a critical role in response to infection with these viruses.26 The important role of NLRP3 has also been indicated and here signalling vis this molecule results in activation of pro-inflammatory cytokines such as IL-1β and IL-18.13,25,34
In this study, among 14 cytokines analysed the significant increase in the levels of IL-1β, IL-2, IL-4, IL-6, IL-8, IL-12, IL-15, TNF-a, and IFN-γ were observed in samples of Flu A, H1N1, RV + H1N1, FluB + H1N1, RV, RSV, RV + RSV + EV with single-stranded RNA, and HAdV with double-stranded DNA. This result corresponds with a previous study showing the recognition of single-stranded RNA viruses by TLR3/8 and double-stranded DNA by TLR 7/9 which resulted in activation of type I IFN and inflammatory cytokines.35 Type I IFN production is one of the key cytokines that directs T cell responses toward Th1-type and cell-mediated immunity.36 In the present study, respiratory viral infection due to FluB+H1N1was associated with significantly higher concentrations of IL-1β, TNF-a, IL-8, IFN-γ when compared with other viruses. It has been documented that influenza virus elicited higher levels of INF-γ, IL-4, and IL-2 compared to human metapneumovirus and RSV in infants.28 Studies on influenza virus and RSV have shown that the differential induction shown is due to the involvement of plasmacytoid dendritic cells (pDCs) at a lower level in the RSV-infected animal.21 However, in this study rhino virus infection showed the lowest expression level of IL-2, IL-4 IL-6, IL-8, TNF-a, IFN-γ and GS-CSF as compared to other viruses.
Interestingly, our results showed significant decrease in level of IL-10 in samples of Flu A, Flu H1N1, FluA + H1N1, RV + H1N1, FluB + H1N1, RV + RSV + EV and HAdV. IL-10’s directly affects T and NK cell function via monocyte-macrophages which prevents expression of MHC class II and costimulatory molecule B7-1/B7-2 on these cells, thus restricting the production of some cytokines such as IL-6, IL-18, TNF-a, IL-1a and β and IL-12. IL-10 also controls the CD4 T cell activity and inhibits proliferation and production of IL-2, IFN-γ, IL-4, IL-5 and TNF-a. Thus IL-10 controls innate and adaptive Th1 and Th2 reactions and this occurs via limiting activation and differentiation of T cells in the lymph nodes apart from suppressing pro-inflammatory responses in tissues.23,24 Downregulation of IL-10 in our results and upregulation of IL-2, IL-4, IL-12, IFN-γ and TNF-a suggested that innate and adaptive Th1 and Th2 responses has been activated against studied respiratory viruses. Additionally, higher expression of GM-CSF was observed in all groups especially in patients with Flu B and H1N1. It has been investigated that GM-CSF is an essential factor to provide the protection against infection with some respiratory viruses such as influenza and RSV.
Activation of cytokines involves the comprehensive host immune responses against viral infection. One should not rule out the possibilities of having different level of cytokines in the host once the virus has been cleared. Therefore, future studies should consider having two time points (during and post infection) of serum cytokine assessment in order determine signature cytokines that are raised only in patients who are positive for RNA of the viruses.11 The present study, also investigated programmed cell death in infected host cells. The effect of the different respiratory viruses on the receptors for the TNF related apoptosis inducing ligand (TRAILR) indicated significant upregulated levels of TRAIL-R1 and TRAIL-R2 in all virus detected samples. It has been reported that up-regulation of TRAIL-R1 and TRAIL-R2 leads to induction of apoptosis. However, over expressions of TRAIL-R3 and TRAIL-R4 inhibit the TRIAL. In this study, no significant upregulated or downregulated expression levels of TRAIL-R3 and TRAIL-R4 was observed in all samples showing the apoptosis may have activated through TRAILR receptors. TRAILR-3 prevents the formation of DISC (TRAIL-R2 associated death inducing signalling complex) while TRAIL-R4 binds to TRAIL-R2 DISC to prevent caspase recruitment within DISC and activating NF-kB to inhibit TRAIL induced apoptosis.
Various pro-apoptotic and anti-apoptotic proteins up-regulated in samples including Flu A, H1N1, RV + H1N1 + Flu A + H1N1, Flu B + H1N1, RV, RSV, RV + RSV + EV, HAdV and RV + H1N1. Pro-apoptotic bcl-2 markers such as BAX, BAD and BIM, CD40, CD40L, Caspase 3 and Caspase 8, P21 and p53 was most frequent among studied groups compared to healthy individuals. Pro-apoptotic markers of the bcl-2 family induce the caspase activation through the caspase release from the death antagonists and this prompts release of mitochondrial apoptogenic factors into the cytoplasm. Up-regulation of Caspase 3 and caspase 8 in virus detected samples imply that caspase 8- dependent activation of caspase 3 leads to induction of apoptosis. Significantly, up-regulated expression level of CD40 and CD40L is able to induce apoptosis through caspase-depended pathways or activation of nuclear factor-κB (NF-κB), AP-1 and CD95.
Of anti-apoptotic proteins, HSP27, sTNF-R1, sTNF-R2, IGF-II, were upregulated in some virus detected samples including Flu A, H1N1, Flu A + H1N1, Flu B+ H1N1, RV, RSV, RV + RSV + EV, HAdV and RV + H1N1. Upregulated of these anti-apoptotic proteins indicates that the viruses might modulate host cell apoptotic responses to supress host immune system. We also detected several down-regulations in anti- apoptotic proteins in virus detected samples. Among the down-regulated proteins, bcl-2 and bcl-w showed the highest fold-differences compared with controls in samples including Flu A, H1N1, RV + H1N1, Flu A + H1N1, Flu B+ H1N1, PIV3 + PIV4, HboV + HCoV-HKU1 + HCoV-229E, and HAdV. Anti-apoptotic bcl-2 markers prevent apoptosis in the mitochondria through inhibition of release of apoptogenic factors, such as cytochrome c or by deactivation of caspases which is through their proficiencies to sequester pro-caspases. The other down-regulated proteins were HSP60, HSP70, livin, Survivin, IGF-I, IGF-1sR, XIAP and cIAP-2. Heat shock proteins (HSPs) bind to apoptosis protease-activating factor 1 (Apaf-1) disrupting apoptosome formation. It has been shown that viruses are able to activate HSPs to reduce apoptosis, but later, control these proteins to induce apoptosis for effective viral dissemination. Survivin, Livin, XIAP and cIAP-2 are members of inhibitor of apoptosis proteins (IAP) that exert their anti-apoptotic activity through binding to caspase 3, 7 and 9 as the effector caspases in the signalling pathway of apoptosis. IGF-I and IGF-II can activate insulin-like growth factor type I (IGF-I) receptor (IGF-IR) to introduce several signal transduction pathways that facilitate apoptosis suppression. Downregulation of these anti-apoptotic proteins suggesting that immune response in studied individuals had a protective role in inhibiting the virus to complete its replication and to disseminate infectious progeny viruses., while down-regulation of some pro-apoptotic markers such as p21, p27, BAX, CD40 or IGFBPs shows that these respiratory viruses have the capacity to disable the host cell apoptotic mechanisms that may be obligatory for viral life cycle completion.
In our study, peripheral blood samples were used to determine adaptive immune responses to a respiratory infection. The main limitation in this study was the sample size. Despite the major limitation of small sample size, this study showed significant alterations in different types of genes and cytokines including proinflammatory, Th1 and Th2 accompanied with the over expression of pro-apoptotic proteins suggesting the immune system can prevent the virus from completing the replication cycle and in producing infectious progeny. However significant up-regulation of some anti-apoptotic genes or down regulation of some pro-apoptotic proteins shows that respiratory viruses have evolved strategies to subvert the immune system. It is this balance between viral replication and control by the immune system that determines the pathogenicity and virulence of the virus. A better understanding of the molecular mechanisms of how viruses interfere with innate immunity and programmed cell death might contribute to the design of new therapeutic tools to treat or prevent respiratory viral infections. This study opens up investigations including identification of biomarkers that may enable therapeutic approaches to viral respiratory infections especially FluA, RV and RSV.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SDS conceptualized project, validated results, reviewed and edited paper. RM curated data and wrote original draft of paper. SMW curated data and wrote original draft. CSR obtained funding, administered project and resources. YKY designed methodology and obtained software and drafted paper. AB performed investigations.
FUNDING
This study was supported by the University of Malaya Research Grant (UMRG-PR021B-13HTM).
ETHICS STATEMENT
The study was approved by Scientific and Ethical Committee of University of Malaya Medical Centre (UMMC), Malaysia, (Ethics Committee/ IRB Reference No: 20143-60).
AVAILABILITY OF DATA
All datasets generated or analysed during this study are included in the manuscript.
- Aponte FE, Taboada B, Espinoza MA, et al. Rhinovirus is an important pathogen in upper and lower respiratory tract infections in Mexican children. Virol J. 2015;12(1):31.
Crossref - Bahadoran A, Lee SH, Wang SM, et al. Immune responses to influenza virus and its correlation to age and inherited factors. Front Microbiol. 2016;7:1841.
Crossref - Bhoj VG, Sun Q, Bhoj EJ, et al. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008;105(37):14046-14051.
Crossref - Bustelo XR. Regulatory and signalling properties of the Vav family. Mol Cell Biol. 2000;20(5):1461-1477.
Crossref - Dong Z, Davidson D, Perez-Quintero LA, Kurosaki T, Swat W, Veillette A. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 2012;36(6):974-985.
Crossref - Elgizouli M, Logan C, Grychtol R, Rothenbacher D, Nieters A, Heinzmann A. Reduced PRF1 enhancer methylation in children with a history of severe RSV bronchiolitis in infancy: an association study. BMC Pediatr. 2017;17(1):65.
Crossref - Erf G. Cell-mediated immunity in poultry. Poult Sci J. 2004:83(4):580-590.
Crossref - Galandrini R, Palmieri G, Piccoli M, Frati L, Santoni A. Role for the Rac1 exchange factor Vav in the signaling pathways leading to NK cell cytotoxicity. J Immunol. 1999;162(6):3148-3152.
- Gavala ML, Bertics PJ, Gern JE. Rhinoviruses, allergic inflammation, and asthma. Immunol Rev. 2011;242(1):69-90.
Crossref - Newton AH, Cardani A, Braciale TJ. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol. 2016;38(4):471-482.
Crossref - Kobayashi SD, Braughton KR, Whitney AR, et al. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. PNAS. 2003;100(19):10948-10953.
Crossref - Aziz H, Zaas A, Ginsburg GS. Peripheral blood gene expression profiling for cardiovascular disease assessment. Genom Med. 2007;1(3-4):105-112.
Crossref - Simmons CP, Popper S, Dolocek C, et al. Patterns of Host Genome-Wide Gene Transcript Abundance in the Peripheral Blood of Patients with Acute Dengue Hemorrhagic Fever. J Infect Dis. 2007;195(8):1097-1107.
Crossref - Meadows SK, Dressman HK, Muramoto GG, et al. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One. 2008;3(4):e1912.
Crossref - Robinson JL, Lee BE, Kothapalli S, Craig WR, Fox JD. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis. 2008;46(7):e61-e64.
Crossref - Palmer C, Diehn M, Alizadeh AA, Brown, PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115.
Crossref - Anselmo LM, Vilar FC, Lima JE, Yamamoto AY, Bollela VR, Takayanagui OM. Usefulness and limitations of polymerase chain reaction in the etiologic diagnosis of neurotoxoplasmosis in immunocompromised patients. J Neurol Sci. 2014;346(1-2):231-234.
Crossref - Woods CW, McClain MT, Chen M, et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS One. 2013;8(1):e52198.
Crossref - Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6(3):207-217.
Crossref - Whitmire JK. Induction and function of virus-specific CD4+ T cell responses. Virol. 2011;411(2):216-228.
Crossref - Weil R, Israel A. T-cell-receptor-and B-cell-receptor-mediated activation of NF-κB in lymphocytes. Curr Opin Immunol. 2004;16(3):374-381.
Crossref - Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23: 601-649.
Crossref - Wortzman ME. The Role of TNFR Family Members, TNFR2 and CD30, in the T cell Response Against Viral Infection (Thesis). 2014. https://tspace.library.utoronto.ca/handle/1807/72595
- Xu AL. Amphioxus Immunity: Tracing the Origins of Human Immunity. Academic Press, United Kingdom. 2015.
- Stern LJ, Brown JH, Jardetzky TS, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368(6468):215-221.
Crossref - Shiina T, Inoko H, Kulski J. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64(6):631-649.
Crossref - Montaldo E, Vitale C, Cottalasso F, et al. Human NK cells at early stages of differentiation produce CXCL8 and express CD161 molecule that functions as an activating receptor. Blood. 2012;119(17):3987-3996.
Crossref - Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA-E through mostly shared but partly distinct sets of HLA-E residues. Eur J Immunol. 2004;34(1):81-90.
Crossref - Masilamani M, Nguyen C, Kabat J, Borrego F, Coligan JE. CD94/NKG2A inhibits NK cell activation by disrupting the actin network at the immunological synapse. J Immunol. 2006;177(6):3590-3596.
Crossref - Emuzyte R, Firantiene R, Petraityte R, Sasnauskas K. Human rhinoviruses, allergy, and asthma: a clinical approach. Medicina (Kaunas). 2009;45(11):839-847.
Crossref - Jartti T, Kuusipalo H, Vuorinen T, et al. Allergic sensitization is associated with rhinovirus-, but not other virus-, induced wheezing in children. Pediatr Allergy Immunol. 2010;21(7):1008-1014.
Crossref - Kreijtz J, Fouchier R, Rimmelzwaan G. Immune responses to influenza virus infection. Virus Res. 2011;162(1):19-30.
Crossref - Noble A. Molecular signals and genetic reprogramming in peripheral T-cell differentiation. Immunology. 2000;101(3):289-299.
Crossref - Tamura SI, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57(6):236-247.
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1-20.
Crossref - van de Sandt CE, Kreijtz, JH, Rimmelzwaan GF. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses. 2012;4(9):1438-1476.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.