ISSN: 0973-7510
E-ISSN: 2581-690X
The microbiome, a community of microorganisms in the body, is currently used as a biomarker in many disease prognoses. Prevotella, Turicibacte, Bacteroides, Firmicutes/Bacteroidetes are frequently used as a biomarker for rheumatoid arthritis, colorectal cancer, and obesity in ordered. The amount of gut microbiota can be changed depending on various factors such as diet, lifestyle, and exercise. However, there is unclear on how the exercise is really effective to be a disease prevention. The present study aims to investigate the different exercise intensities on gut microbiome abundance changes that could be used as a disease biomarker. Eighteen Sprague-Dawley rats were arranged (n=6 per group) into 3 exercise intensity levels on treadmills including non-exercise group, high -exercise group (20 – 25 m/min for 60 min), and light-exercise group (10 – 15 m/min for 60 min). Rats were weighted every 2 days and stools were collected and preserved in DNA/RNA shield each week. The bacterial 16S rDNA of microbiome in feces samples was sequenced and analyzed. After week eighth of the interventions, from operational taxonomic unit (OTUs) abundance, we found that the relative abundance in bacterial genera in Prevotella and Firmicutes/Bacteroidetes were significantly correlated with the experiment timepoints in different exercise intensities (Pearson’s correlation, P<0.05) compare to other genera. The exercise intensities and exercise durations can affect the relative abundance in the bacteria genus which the abundance genus Prevotella and Firmicutes/Bacteroidetes could be used as a new standard biomarker in exercise as a disease prevention and exercise prescriptions. From the funding limitations, we could conclude the research results based on our data and statistic. Future research should utilize a longer investigation period.
Aerobic exercise, Firmicutes/Bacteroidetes, Prevotella, Turicibacter
The gut microbiome comprises trillions of bacteria, fungi and other microorganisms in the gastrointestinal (GI) tracts of humans and animals.1 This microbiome is composed of bacteria, archaea, viruses and eukaryotic microbes impact health and also induce disease.2 Several factors including lifestyle, physical activity and diet affect the relative abundance and diversity of the gut microbiome. Physical activity increases the diversity of gut microbial populations, with higher relative abundances in professional athletes than the general population.1 Gut microbiome relative abundance was higher, with more diversity in elite professional rugby players than the general population. Higher diversity of gut microbiota promotes health and impacts amino acid biochemical pathways, antibiotic biosynthesis, carbohydrate metabolism and fecal metabolite levels that all enhance muscle turnover and overall health outcome.3,4 Relative microbiome changes in diversity after 14 weeks of exercise intervention were investigated in 18 obese and 14 lean individuals,5 while a six-week exercise intervention found changes in microbiome taxa.6
Recently, various microbiomes such as Prevotella, Turicibacter and Bacteroides have been used as biomarkers after exercise intervention treatments for rheumatoid arthritis (RA), colorectal cancer (CRC) and obesity (OB).7
Turicibacter, a gram-positive non-spore-forming bacterium with irregularly misshapen rods, is most commonly used as a colorectal cancer (CRC) marker in humans and animals. Turcibacter is one million-fold more abundant in the colon than in the small intestine.7 These types of bacteria show reduced or increased relative abundances depending on diet and physical activity.8,9 A study in C57BL/6 female mice found that Turicibacter increased in the mice study group that consumed a high-fat diet, which can lead to colorectal cancer (CRC),8 while a recent study showed that African Americans who consumed a high-fat diet for 2 weeks showed decreased bacterial diversity and short chain fatty acids (SCFAs) compared to those who consumed a high fiber and low-fat diet.3,10 Exercise was found to impact Turicibacter relative abundance.11,12 A study in mice found that different exercise intensities significantly reduced numbers of Turicibacter in mice feces,11,12 while premenopausal women aged 18-40 had higher relative abundances of Turicibacter in the sedentary group compared to the active group.9 Turicibacter population depends on host immune cell systems and Turicibacter can be abolished when the host immune is deficient.13 Dimitriu et al. found that deficiency of CD45, B and T cells in an immunodeficiency mouse model completely destroyed Turicibacter population in the GI tract compared with wild-type mouse,14 while Turicibacter showed a bidirectional relationship between the mammalian immune system and pathogenesis of intestinal bowel disease (IBD).13 Current studies also investigated colonic pathogenies that occurred when the bacterial community increased, with slow growth in SCFAs after damage to the colonic mucus by high fat accumulation in physically inactive people.3,10 However, the mechanisms of how colorectal cancer occurs from bacteria remain unclear, and further research on exercise intervention should focus on the relative abundance of Turicibacter.
Inactive physical exercise can cause body weight increase in individual. Bacteroidetes/Firmicutes are gram-negative anaerobic bacteria which have high relative abundance in obesity and physical leanness in mammals.15 Bacteroidetes/Firmicutes are significantly associated with weight gain and high BMI.15 A study reported that Bacteroidetes/Firmicutes inhibit obesity by hydrolyzing carbohydrates that are indigestible in the gut to produce SCFA with protein receptors such as 3/G protein-coupled receptor 41 (FFAR3/GPR41), causing reduced food consumption and promoting higher hormone leptin levels to inhibit obesity.16,17 By contrast, another study suggested that increased Firmicutes and Bacteroidetes did not correlate with SCFA production. Bacteroidetes produced acetate and propionate in obese individuals, while Firmicutes produced butyrate that impacted health and regulated energy metabolism and hormone gene expression to induce leptin.16,18 Moreover, Firmicutes also affect the chemical compound propionate in the colon that stimulates GLP-1 and PYY release by L-entero-endocrine and reduces appetite.19 Furthermore, a study of how physical activity and diet impacted Bacteroidetes/Firmicutes found an inverse correlation between these two phyla. Bacteroidetes reduced in relative abundance after a high-fat diet but increased in relative abundance after exercise, while Firmicutes reduced in relative abundance after both exercise and a high-fat diet. However, one study found that Firmicutes increased in relative abundance after exercise, while Bacteroides decreased.20 Exercise can change the relative abundance of Bacteroidetes/Firmicutes is still in current controversies in many studies which there are needed to be further observed.
Another bacteria that has been known for the rheumatoid arthritis marker is Prevotella. The bacteria relate to many complicated factors such as exercise, T-cells, cytokines network, dendritic cells and macrophages. Those factors can be stimulated by Prevotella to induce inflammatory and autoimmune disorders.21-23 Moreover, one study found Prevotella affected high-level expressions of IL-6, IL-23 and Th17 that can cause rheumatoid arthritis,24 while amounts of bacteria were also associated with carbohydrate and SCFA structures.15 Interestingly, Prevotella induced arthritis through the differentiation of CD4+ T lymphocytes to the Th17 subset in rats,24-26 while Japanese and European rheumatoid arthritis patients exhibited higher 16S rRNA genes of Prevotella than American patients.24 A study in db/db mice found that low-intensity treadmill exercise at 5 days per week for 6 weeks showed non-significant Prevotella relative abundance in both the control and interventional groups,20 while professional football players recorded higher Prevotella relative abundance than sedentary individuals.27 Numerous assumptions have been made concerning how gut microbiota alter body systems related to varied factors including bacterial diversity, bacterial family, environments, lifestyles and internal conditions. This study aims to evaluated the effects of high-intensity exercise on the gut- microbiome with confident the high-intensity exercise decrease the bacterial biomarkers relative abundance.
Animals
Eight to eleven weeks old male Sprague-Dawley rats were purchased from Nomura Siam International and housed at Chulalongkorn University Laboratory Animal Center (CULAC). All animals were assigned into 6 cages, 3 animals per cage, and maintained under a 12/12 light-dark cycle at 22°C with food and water Ad libitum feeding.
Exercise intervention
After 2 weeks of being quarantined and another week of treadmill training preparation, all animals were divided into 3 groups (6 rats per group). The first group was assigned to receive a light exercise with a low-speed treadmill (10 – 15 m/min) for 60 min. The second group was assigned to receive a heavy exercise with a high-speed treadmill (20 – 25 m/min) for 60 min. The last group was conducted as a control non-exercise group (group 3). The weight of all animals was measured every 2 days. Animals in the exercise groups were allowed to rest for 15 minutes when they show signs of fatigue (stay at the end of the treadmill and be unable to run with sound or electrical stimulation). On day 45, all rats were euthanized by the inhalation of CO2.
Sample collection
Fecal samples from each group were collected every week (on Monday morning) during this study. Samples were harvested and preserved in DNA / RNA shield solution (Zymo Research, USA), then stored at -80°C until performing a further experiment.
DNA isolation, preparation, and sequencing
The fecal suspension was centrifuged at 15,000 rpm for 10 min. The bacterial and fecal debris pellet was lysed by a lysis buffer from the GenUP gDNA extraction kit (Biotechrabbit, Germany). Subsequently, the total DNA was extracted following the directions provided by the manufacturer of the extraction kit.
The V3-V4 region of the bacterial 16S rDNA was amplified using the following primers (341F:5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG3’ and 807R:5’GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3’). The 25 µl PCR reaction comprised 12.5 ng of DNA 1 µM of each primer, and 2×KAPA HiFi HotStart Ready Mix (Roche, USA). The PCR amplification was performed as following conditions; 1 cycle of 98°C for 30 sec; followed by 25 cycles of 98°C for 10 sec, 55°C for 25 seconds, and 72°C for 25 sec; and 1 cycle of 72°C for 10 min, then the reaction was held at 4°C. Afterward, the PCR products were attached with indexes and Illumina sequencing adapter using Nextera XT index kit (Illumina, USA). The amplified PCR product (approximately 550 bp) was clean up using AMPure XP beads (Beckman Coulter, USA). The library was normalized, pooled, and sequenced by paired-end (2×300) on the MiSeq system using V3 reagents according to the Humanizing Genomics Macrogen service.
Data analysis and statistical test
The FASTQ sequences were demultiplexed using MiSeq reporter software (version 2.6.2.3). Subsequently, the sequences were fully processed by using a QIIME2 pipeline (version 2021.4). The paired-end sequences were merged and then filtered based on the Phred quality score (Q20). Afterward, filtered reads were deduplicated and clustered with 97% similarity by VSEARCH.28 The chimeric reads were also filtered out by the UCHIME algorithm.29 Finally, these passed reads were classified by comparing the sequence against the Greengenes Database version 13.5 using the VSEARCH algorithm. The alpha and beta diversity were analyzed based on the implemented plugin of QIIME2.30
The statistical test of alpha diversity between microbiota among different timepoints was t-test (P<0.05). The difference between bacterial communities among exercise groups in 3 timepoints (timepoint 1,5 and 9) determined by beta-diversity was statistically tested by permutational multivariate analysis of variance (PERMANOVA) with P<0.05. The Firmicutes/Bacteroidetes ratio and selected OTUs, which may have potential functions related to the current experiment, including Prevotella, Bacteroides, and Turicibacter, were investigated for their association with 3 different timepoints using Pearson’s correlation analysis (P<0.05).
Sequence output and microbiome diversity
The sequencing output analyzed and classified by QIIIME2 reached the minimum of 20,000 reads per sample. The operational taxonomic unit (OTUs) identified from the sequencing were rarified in rarefaction analysis (as shown in Supplementary Fig. 1). The alpha diversity of microbiome was evaluated by Chao 1 index and Shannon index (Fig. 1a-b). The results showed that Chao 1 richness of light and heavy exercise groups were significantly different (t-test, P<0.05) while the Shannon index showed the significant differences between timepoints 1 and 5 in non-exercise group. The beta diversity based on the Jaccard dissimilarity was analyzed and shown in Fig. 1c. The beta diversity indicated that bacterial microbiome among the exercise group were not significantly different. However, they presented the distinct characteristics in different timepoints (P<0.05, PERMANOVA test)
Fig. 1. Alpha and beta diversity of mice fecal microbiome among 3 time points. The box plot shows the alpha diversity calculated by the (A) Chao1 index and (B) Shannon index. The differences in alpha diversity between group were tested statistically by the t-test (*P<0.05). The beta diversity was evaluated by the Bray-Curtis dissimilarity index and visualized as a (C) principal coordinate analysis plot. The difference in community among time points was tested by PERMANOVA with P<0.05.
Bacterial relative abundance
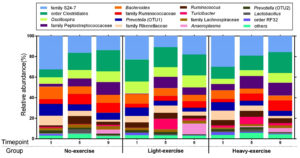
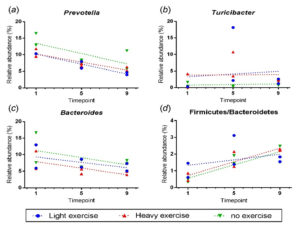
The top 15 bacterial genus levels from all bacteria are shown in the form of relative abundance percentage correlated with various exercise intensities and timepoints shown in Fig. 2. The relative abundance of four bacterial biomarkers had been used for correlation analysis against exercise intensities. The Pearson’s correlation coefficient (r) and P-value were showed in Table 1. Correlation between the time points and these four bacterial biomarkers was also investigated Fig. 3.
Table (1):
The Pearson’s correlation coefficient (r) and significance of correlation (P < 0.05) in each exercise group.
| Bacteria | Light exercise | Heavy exercise | No exercise | |||
|---|---|---|---|---|---|---|
| P | r | P | r | P | r | |
| Prevotella | 0.002* | -0.967 | 0.015* | -0.899 | 0.119 | -0.704 |
| Turicibacter | 0.841 | 0.107 | 0.969 | 0.0209 | 0.677 | 0.219 |
| Bacteroides | 0.305 | -0.507 | 0.144 | -0.672 | 0.303 | -0.051 |
| Firmicutes/Bacteroidetes | 0.485 | 0.359 | 0.013* | 0.905 | 0.028* | 0.859 |
(*P < 0.05) significance in exercise intensity groups correlate with bacteria genera expressions
Fig. 2. Relative abundance of the top 15 most abundant bacterial OTUs identified in fecal microbiome using Qiime2 pipeline. The stacked bar plot shows the average abundance of fecal microbiota from 3 groups of mice (Light exercise, Heavy exercise and No- exercise), which were investigated for 3 timepoints at timepoints 1,5, and 9 of the experiment.
Fig. 3. Correlation between the timepoints and fecal bacterial contents. (A) Prevotella. (B) Turicibacter (C) Bacteroides group (D) Firmicutes/Bacteroidetes.
Table 1 shows the Prevotella has significance (P<0.05) in light- and heavy- exercise groups rather than no-exercise group. Meanwhile, Firmicutes/Bacteroidetes also has significance (P<0.05) in heavy and no-exercise groups compared to three other bacterial genera. The results from Table 1 and Fig. 3 regarding Prevotella which indicates a rheumatoid arthritis disease decrease in numbers in light- exercise (P = 0.002, r = -0.967) and heavy exercise groups (P = 0.015, r = -0.899), which shows that the exercise durations affect this type of bacteria. Also, Firmicutes/Bacteroidetes genus which are an obesity biomarker increases in heavy exercise group (P = 0.013, r = 0.905) and no-exercise groups (P = 0.028, r =0.859). There both show the heavy and no-exercise groups have the least effect on this type of bacteria impact to obesity. In the meantime, other candidate bacteria are not significant (P < 0.05).
The scatter plots showed the relative abundance (%) of within 3 groups of rats (light- exercise, heavy- exercise and no- exercise) against 3 timepoints (Timepoints 1, 5, 9) following the collection dates from protocols. The dashed line represents the estimation line from the regression analysis. Pearson’s correlation test was carried out to present the correlation (r) and the significance of such correlation (P<0.05).
Our findings show that aerobic exercise affects the microbiome relative abundance percentage. As with Prevotella, this shows that the relative abundance slightly decreased in both the heavy and light exercise intensity groups when compared with the non-exercise group. This finding indicates that the non-exercise group has a higher relative abundance, which is associated with higher risk in rheumatoid arthritis prognosis.31,32 This could be related to the study duration, but could also be because Prevotella includes various species with a high degree of genetic diversity within the population. This genetic diversity could be associated with the relative abundance of the results. Prevotella also have different characteristics, despite consuming the same diet, having the same environment, and inheriting the same phylogenetic characteristics.33,34 Meanwhile, different Prevotella species can also result in the occurrence of varied diseases among humans and animals, including rheumatoid arthritis.
Another possibility is that some Prevotella species can change the form of Th17 to initiate unbalancing of T helper type 17 (Th17) cells and T regulatory (Treg) cells, which is linked to high rheumatoid arthritis prognosis. This novel approach requires further investigation.35,14
Firmicutes/Bacteroidetes relate to short-chain fatty acid propionate, acetate, and butyrate, which are factors for obesity and energy use. From the Figs., rat genotype differences can stimulate energy breakdown and metabolism varies for each rat.36 Firmicutes/Bacteroidetes have diverse genetics that can induce many diseases, but this depends on the host cell gut of the individual.28 Another possibility that resulted in Firmicutes/Bacteroidetes being significant in the heavy exercise group (Table 1 and Fig. 3) was because they have their own outer membrane vesicles (OMVs) which could help Firmicutes/Bacteroidetes adapt themselves to diverse niches environments which could give Firmicutes/Bacteroidetes be significant related to obesity and energy used.37 In the future research we aim to utilize a longer investigation period for the study.
Exercise intensity differences can change the relative abundance percentages of bacteria, but they depend on the timepoint and exercise intensities. The microbiome in the gut in addition to both external and internal factors can influence bacterial abundance, which could be used as disease biomarkers to prove the effectiveness of exercise on disease prevention and exercise prescriptions. Finally, gut microbiome mechanisms require further future investigation.
ACKNOWLEDGMENTS
The authors would like to acknowledge and offer appreciation to the Chulalongkorn University Laboratory Animal Center (CULAC) and the Laboratory Research Assistance Staffs for facilitating our rodents during the research period in animal part.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
VT, SP, WS, TP organized research ideas.PC performed DNA isolation, preparation, and sequencing. VS, VT, WS performed the data analysis, statistical test and wrote the data results. VT drafted the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by the TINARATHPATRA Co., Ltd with grant ID 46/158S1.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Chulalongkorn University Animal Care, Thailand with protocol number 2073010.
AVAILABILITY OF DATA
The data generated during the current study are available in the SRA database repository, Accession No. SRR17687173-17687190.
- Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7(1):17-44.
Crossref - Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr opin Gastroenterol. 2015;31(1):69-75.
Crossref - Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol. 2019;17(2):275-289.
Crossref - Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913-1920.
Crossref - Allen JM, Mailing LJ, Niemiro GM, et al. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc. 2018;50(4):747-757.
Crossref - Munukka E, Ahtiainen JP, Puigbo P, et al. Six-Week Endurance Exercise Alters Gut Metagenome That Is not Reflected in Systemic Metabolism in Over-weight Women. Front Microbiol. 2018;9:2323-2323.
Crossref - Gagniere J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22(2):501-518.
Crossref - Yang J, McDowell A, Kim EK, et al. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp Mol Med. 2019;51(10):1-15.
Crossref - Bressa C, BailEn-Andrino M, PErez-Santiago J, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PloS one. 2017;12(2):e0171352.
Crossref - O’Keefe SJ, Li JV, Lahti LM, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6(1):6342.
Crossref - Evans CC, LePard KJ, Kwak JW, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PloS One. 2014;9(3):e92193.
Crossref - Cook MD, Allen JM, Pence BD, et al. Exercise and gut immune function: evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol Cell Biol. 2016;94(2):158-163.
Crossref - Allen JM, Berg Miller ME, Pence BD, et al. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol. 2015;118(8):1059-1066.
Crossref - Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ Microbiol Reports. 2013;5(2):200-210.
Crossref - Indiani CMdSP, Rizzardi KF, Castelo PM, Ferraz LFC, Darrieux M, Parisotto TM. Childhood Obesity and Firmicutes/Bacteroidetes Ratio in the Gut Microbiota: A Systematic Review. Childh Obes. 2018;14(8):501-509.
Crossref - Duan M, Wang Y, Zhang Q, Zou R, Guo M, Zheng H. Characteristics of gut microbiota in people with obesity. PloS One. 2021;16(8):e0255446.
Crossref - Ohira H, Tsutsui W, Fujioka Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J Atheroscler Thromb. 2017;24(7):660-672.
Crossref - Soliman MM, Ahmed MM, Salah-Eldin A-E, Abdel-Aal AA-A. Butyrate regulates leptin expression through different signaling pathways in adipocytes. J Vet Sci. 2011;12(4):319-323.
Crossref - Chambers ES, Viardot A, Psichas A, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744-1754.
Crossref - Lambert JE, Myslicki JP, Bomhof MR, Belke DD, Shearer J, Reimer RA. Exercise training modifies gut microbiota in normal and diabetic mice. Appl Physiol Nutr Metab. 2015;40(7):749-752.
Crossref - Bailey CF. The treatment of chronic rheumatic and rheumatoid arthritis by radiant heat and cataphoresis. BMJ. 1909;1(2505):13-15.
Crossref - Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202.
Crossref - Pianta A, Arvikar S, Strle K, et al. Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2017;69(5):964-975.
Crossref - Maeda Y, Takeda K. Role of Gut Microbiota in Rheumatoid Arthritis. J Clin Med. 2017;6(6):60.
Crossref - Drago L. Prevotella Copri and Microbiota in Rheumatoid Arthritis: Fully Convincing Evidence? J Clin Med. 2019;8(11):1837.
Crossref - Rogier RL, Ederveen TH, Boekhorst J, et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome. 2017;5(1):63.
Crossref - Celik A. The Relationship Between Provetella Genus in Intestinal Flora and Exercise and Diet Styles of Professional Football Players. Progress in Nutrition. 2021;23(3):e2021103.
Crossref - Liang D, Leung RK-K, Guan W, Au WW. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathogens. 2018;10(1):3.
Crossref - Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194-2200.
Crossref - Desantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069-5072.
Crossref - Bycura D, Santos A, Shiffer A, et al. Impact Of Different Exercise Modalities On The Human Gut Microbiome: 676. Medicine and Science in Sports and Exercise. 2021;53(8S):230-230.
Crossref - Bermon S, Petriz B, Kajeniene A, Prestes J, Castell L, Franco OL. The microbiota: an exercise immunology perspective. Exerc Immunol Rev. 2015;21:70-79.
- Guerreiro CS, Calado A, Sousa J, Fonseca JE. Diet, Microbiota, and Gut Permeability-The Unknown Triad in Rheumatoid Arthritis. Front Med. 2018;5:349.
Crossref - Bischoff SC, Barbara G, Buurman W, et al. Intestinal permeability–a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14(1):189.
Crossref - Moreno J. Prevotella copri and the microbial pathogenesis of rheumatoid arthritis. Reumatologia clinica (Barcelona). 2015;11(2):61-63.
Crossref - Zhong Y, Nyman M, Fak F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res. 2015;59(10):2066-2076.
Crossref - Tafti ZSM, Moshiri A, Ettehad-Marvasti F, et al. The effect of saturated and unsaturated fatty acids on the production of outer membrane vesicles from Bacteroides fragilis and Bacteroides thetaiotaomicron. Gastroenterol Hepatol Bed Bench. 2019;12(2):155-162. PMCID: PMC6536021
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.