ISSN: 0973-7510
E-ISSN: 2581-690X
COVID-19 detection via lateral flow antigen assays (LFA) are rapid and economically acquiescent to infrastructure facile healthcare settings. Early, prompt identification of cases to facilitate patient isolation and supportive management is the essence of rapid diagnostic tests. Given the backdrop of post COVID-19 pandemic-molecular testing still remains a costly affair. Additionally, molecular assays are incapable of distinguishing remnant RNA from replication competent viruses. In this scenario, we explore the diagnostic consonance of SARS-CoV-2 LFAs with RT-PCR cycle threshold, in a likelihood that it could be used as a surrogate marker for infection transmissibility. Rapid COVID-19 LFA results were compared with Real-time PCR for detection of SARS-CoV-2 in nasopharyngeal swabs. Two hundred rapid antigen positive nasopharyngeal swabs obtained from COVID-19 suspects/contacts/preoperative/screening patients were subjected to RT-PCR to study the correlation with cycle threshold (CT) values obtained for all the antigen positive cases. 200 Rapid COVID-19 LFA positive samples were analyzed in the present study. Amidst the LFA positive samples included in the study 187 (93.5%) were found to have concordant results when subjected to the gold standard Real-time PCR. Discordant results were documented in 13 (6.5%) COVID-19 LFA positive samples which were found to be negative by RT-PCR. The average Cycle threshold values were found to be 23.75 for E gene, 25.36 for N gene and 24.07 for RdRp gene. The average PCR Cycle threshold of LFA positive cases remained significantly undeterred (p<0.5) throughout the time period of the study stipulating the undaunted viral load across the different waves of the pandemic. Maximum association of LFA positivity with symptom-manifestation was seen during the 1st wave of COVID-19 (September-December 2020 in India). The association of symptoms with LFA test positivity reduced to a significant extent during the 3rd wave of the pandemic in January 2022 (p<0.5) indicating the reduced clinical severity but not infectivity of the SARS-CoV-2 infection during the 3rd wave of the pandemic. Lateral flow assay based diagnostic tests are technically & economically convenient modalities with significant interest concordance in comparison with RT-PCR. Definitive advantage in terms of achieving quick patient triage and thereby patient management can be achieved with the use of these tests.
SARS-CoV-2, Lateral Flow Antigen Assay, RT-PCR, Cycle Threshold, COVID-19
Diagnosis forms the cornerstone of the World Health Organization (WHO) Test-Isolate-Trace strategy to break the transmission chain and cease the spread of SARS-CoV-2.1 Each juncture of this strategy is subject to hindrances and uncertainties attributed to time dependent processes. Rapid accurate diagnosis is the linchpin in this strategy. Laboratory physicians yearn for a diagnostic test capable of quick and accurate detection of individuals harboring replication competent virions.
Ever since the inception of the COVID-19 pandemic, molecular methods targeting genomic RNA have been the gold standard primary diagnostic modalities. These molecular diagnostic assays are incapable of differentiating remnant viral RNA shedding from the parole of replication-competent virions. Thus, genotypic diagnostics may not be applicable to predict virus transmissibility. Labour intensive culture-based virus propagation assess infectiousness of the shed virus. However, viral culture techniques are time and resource consuming, therefore remain unsuitable for implementation as a mass testing strategy. Additionally, viral culture facilities are available only in reference laboratories and cannot be done at peripheral laboratories.
At the present juncture of the pandemic, given the perpetual SARS-CoV-2 spread along with the rise in variants of concern (VOC) is resulting in economic clang and infrastructure exhaustion. In seasonal epidemic prone countries COVID can present as a diagnostic dilemma with coexisting infections. Therefore a rapid, cost-effective diagnostic marker for SARS-CoV-2 infectiousness is of unparalleled dominion to combat COVID in resource limited settings of a highly populous country.
The Ct (cycle threshold) value determined by RT-PCR holds promise to serve as a proxy value for infectiousness. After taking into account viral load kinetics Ct value can support infection control decisions.2 Samples with Ct values lower than 35 for nucleocapsid (N) gene and RNA-dependent RNA polymerase gene (RdRp) find positive correlation with infectiousness.2 Patients with a Ct value <30 on molecular assays show 1.5 times higher transmission rates.3 The Present study proposes to look for RT-PCR cycle threshold correlation with positive Rapid COVID-19 LFA results and the ability of LFAs to be used as a proxy, inexpensive marker of infectiousness in mass settings given the present global post pandemic scenario.
The present study is a retrospective cross-sectional hospital based study of 1.25 years duration. The time frame of the study was September 2020-January 2022. The study was conducted in the Department of Microbiology, JSS Hospital, by taking the nasopharyngeal swabs which were sent for RT-PCR for SARS-CoV-2 and COVID-19 antigen detection in suspected cases of COVID-19. Indian Council of Medical Research (ICMR) validated point of care test (POCT) with sensitivity of 84.38% and specificity of 100% was utilized for LFA rapid antigen detection (Standard Q Covid 19 Antigen kit, SD Biosensor). Real-time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) test kit used in our study was an ICMR evaluated kit with a certified sensitivity & specificity of 100%.4 200 patient nasal samples received from various inpatient wards and ICUs of a tertiary care hospital in South India found to be positive by COVID-19 LFA rapid antigen test (RAT) were included in the present study.
SARS-CoV-2 Rapid antigen testing
Nasopharyngeal samples collected from COVID-19 suspected patients5,6 were subjected to COVID-19 antigen testing by a POC. The nasopharyngeal swab was inserted and stirred in the extraction buffer tube. The extraction buffer tube is then capped with a nozzle. Three drops of extracted specimen was applied into the specimen well of the cassette. The results were read and documented within 30 minutes of specimen application.
SARS-CoV-2 RT-PCR testing
Simultaneously nasopharyngeal and oropharyngeal swabs from each COVID-19 suspected patient7 was transported to the molecular microbiology laboratory in a vial of viral transport medium. All samples were subjected to in-vitro RT-PCR assay for the specific detection of SARS-CoV-2 E-gene, N-gene and RdRp-gene and the corresponding Cycle Threshold (Ct) value was determined. RNase P served as the internal control. The test assay used was a single tube assay based on Berlin protocol.8,9 RNA extraction was done by using an IVD approved kit (NucleoDx RT RNA kit). The reaction mix was prepared keeping a sample input volume of 200 µl, the thermal cyclic conditions were set as per manufacturer instructions (NeoDxCoviDxTM mPlex-4R covid 19 RT-PCR Detection Kit). A specimen was considered positive for COVID-19 in case the CT value was < 40 cycles for E gene, N gene and RdRp target. The corresponding CT value thus obtained for each specimen was recorded.
200 Nasopharyngeal samples from the selected patients which were positive by rapid antigen testing and simultaneously tested by RT-PCR assay were included in the present study. The RT-PCR Ct correlation with the POCT LFA positivity was compared. Data thus obtained was scrutinized for statistical significance by p value estimation using ANOVA calculator software.10 P value ≤ 0.5 was considered significant. The demographic details and clinical details were recorded and analysed.
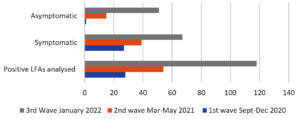
A retrospective cohort of 200 SARS-CoV-2 LFA positive patient samples which had been simultaneously received for COVID RT-PCR testing has been analyzed in the present study. The time line of the samples obtained was docketed based on the COVID-19 pandemic waves in India. Amidst 200 samples a major chunk of 118 samples (59%) were obtained during the 3rd wave in January 2022 (Figure 1).
The mean age of the subjects included in the study was 39.3 years (range 5 to 81 years). The male: female ratio was found to be 137:63 and 68.5% of the study subjects were males. As depicted in Figure 1, the proportion of symptomatic individuals were comparatively higher during the first and second waves as against the symptomatic testing positive during the third wave of the pandemic. A significantly higher proportion of asymptomatic individuals tested positive by LFA test for SARS-CoV-2 during the third wave of the pandemic in January 2022.
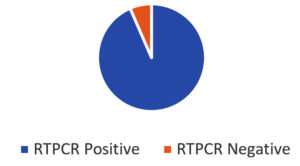
Quantitative RT-PCR performed on the 200 LFA positive samples was analyzed. 187 samples showed concordant results while 13 samples (6.5%) showed discordant results (Figure 2). Majority (10/13) discordant results were recorded during the 3rd wave of the pandemic in January 2022. Amid the 13 discordant results obtained 08 patients were asymptomatic and were screened to obtain surgical clearance and remaining 05 patients had mild symptoms of fever, cough & sore throat.
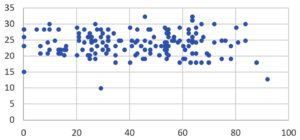
The cycle threshold values for the primer-probe sets targeting envelope (E), nucleocapsid (N) & RNA-dependent RNA polymerase (RdRp) genes showed no significant difference (p>0.5) across the timeline of the captured data. As manifested in Figure 3: The mean Ct value for E gene was 23.75, N gene was 25.36 & RdRp was 24.07 across the timeline of the study. The range of the Ct values obtained in the present study for LFA positive samples was found to be 13-30 for E gene, 17-33 for N gene and 17-33 for RdRp gene. The mean Ct for the 3 genes of interest detected by RT-PCR for each subject with concordant result was analysed against the age of the patients. The Pearson correlation coefficient was found to be -0.0028 showing negative correlation. However, the P value was 0.979 and no significant association of the Ct value across the age groups was noted as depicted in Figure 4.
The wrath of the COVID-19 virus has shaken the world socially, ethically as well as economically. This pandemic brought the diagnostic front to its knees given the high testing load, equipment and taskforce requisites.
The first COVID-19 case in Karnataka, India, was confirmed on March 1st 2020.11 The peak of the first wave in Karnataka was seen between September to December 2020.11 The second wave of cases was documented between March to May 2021. The 3rd surge of COVID cases in South India was seen during January 2022. Present study had documented a significant higher rate of asymptomatic subjects testing positive in the third COVID wave in comparison with the first two waves of the pandemic. A similar finding was documented from western India by Patel et al.12 and eastern India by Singh et al.13 This reduction in the symptom complex can be attributed to two accoutrements. Firstly SARS-CoV-2 being an single stranded RNA virus is prone to higher rate of mutation14,15 and secondly, the successful vaccination coverage across the Indian subcontinent.16
A reliable LFA offers a low cost, accessible, point of care diagnostic facility to rapidly detect subjects harbouring SARS-CoV-2. This rapid diagnostic decision making is of utmost importance to initiate timely intervention for patient welfare as well as to break the public transmission chain. LFAs come with the advantage of minimum potential analytical interference at an affordable cost attributable to the marginal hands on time and nadir technical expertise requirement. The commercially available ICMR approved LFAs are excellent in terms of specificity (95%). However, the sensitivity of the LFA kits are not absolute. Sensitivity of commercially available LFA kits range from 50% upwards,17 thereby opening up the possibility of false negatives. RT-PCR tests come with the advantage of superior analytical sensitivity and specificity. High test sensitivity comes with the risk of increasing the false positives & therefore reducing positive predictive value of the test. Routine RT-PCR for SARS-CoV-2 can be positive in COVID-19 patients for weeks or months following the resolution of their illness1,3; hence, COVID infection control measures will be economically more effective if the diagnostic test performed can distinguish between persistent shedding of non-infectious viral RNA and actively replicating infectious virus. In the present study a concordance of 93.5% was obtained between LFA positives and RT-PCR test result. Similar results were documented in studies by Escriva et al.18
The range of the Ct values obtained in the present study for LFA positive samples was found to be 13-30 for E gene, 17-33 for N gene and 17-33 for RdRp gene. Documented evidence shows inverse relation of Ct values with live virus culture.18 Subjects with high Ct values are unlikely to have infective potential.19 RNA fragments remain detectable in the subjects >14 days post infection; however, the infectious potential substantially reduces after 8 days of infection evident from literature which substantiate that the odds of live virus culture reduced by 33% for every 1 unit increase in the Ct value. The probability of recovering live virus from specimens with Ct value> 35 is very low as per documented evidence.19,20 Considering the above factors SARS-CoV-2 LFAs find utility as proxy infectivity indicating modalities for rapid triaging and patient management.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Goldstein ND, Burstyn I. On the importance of early testing even when imperfect in a pandemic such as COVID-19. Glob Epidemiol. 2020;2:100031.
Crossref - Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483.
Crossref - Al Bayat S, Mundodan J, Hasnain S, et al. Can the cycle threshold (Ct) value of RT-PCR test for SARS CoV2 predict infectivity among close contacts? J Infect Public Health. 2021;14(9):1201-1205.
Crossref - Kumar KR, Mufti SS, Sarathy V, Hazarika D, Naik R. An update on advances in COVID-19 laboratory diagnosis and testing guidelines in India. Front Public Health. 2021;9:568603.
Crossref - Sule WF, Oluwayelu DO. Real-time RT-PCR for COVID-19 diagnosis: challenges and prospects. Pan Afr Med J. 2020;35(Suppl 2):120.
Crossref - Chaimayo C, Kaewnaphan B, Tanlieng N, et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J. 2020;17(1):177.
Crossref - Ministry of Health and Family Welfare (India). Guidelines for notifying COVID19 affected persons by Private Institutes. https://www.mohfw.gov.in/pdf/GuidelinesfornotifyingCOVID19affectedpersonsby PrivateInstitutions.pdf
- Campos KR, Sacchi CT, Goncalves CR, et al. COVID-19 laboratory diagnosis: comparative analysis of different RNA extraction methods for SARS-CoV-2 detection by two amplification protocols. Rev Inst Med Trop Sao Paulo. 2021;63.
Crossref - Said K, de Laurent Z, Omuoyo D, Lewa C, Gicheru E, Cheruiyot R, et al. An optimisation of four SARS-CoV-2 qRT-PCR assays in a Kenyan laboratory to support the national COVID-19 rapid response teams. Wellcome Open Res. 2020;5.
Crossref - Socscistatistics.com. One-Way ANOVA Calculator. http://www.socscistatistics.com/tests/anova/default2.aspx. Accessed June 27, 2023.
- COVID-19, Dashboard. karunadu.karnataka.gov.in. Archived from the original on April 19, 2020.
- Patel AK, Patel D, Shevkani M, et al. COVID-19 patients’ clinical profile and outcome with respect to their vaccination status: A prospective observational multicentre cohort study during third wave in Western India. Indian J Med Microbiol. 2023;41:28-32.
Crossref - Singh KN, Mantri JK. Classifications of COVID-19 Variants Using Rough Set Theory. In: Ambient Intelligence in Health Care 2023. Singapore: Springer; 2023:381-389.
Crossref - Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269.
Crossref - Kaushal N, Gupta Y, Goyal M, Khaiboullina SF, Baranwal M, Verma SC. Mutational frequencies of SARS-CoV-2 genome during the beginning months of the outbreak in USA. Pathogens. 2020;9(7):565.
Crossref - Ministry of Health & Family Welfare (India). India’s cumulative Vaccine coverage. Press Information Bureau, Government of India. Available from: https://pib.gov.in/newsite/pmreleases.aspx?mincode=31 Indian Council of Medical Research (ICMR).
- Advisory on Rapid Antigen Test Kits for COVID-19 (Oropharyngeal swabs / Nasopharyngeal swabs / Oral Saliva) dated: 30.12.2021. Recommended by the National Task Force on COVID-19; 2021. https://www.icmr.gov.in/pdf/covid/kits/archive/List_of_rapid_antigen_kits_30122021.pdf
- Escriva BF, Mochon MD, Gonzalez RM, et al. The effectiveness of rapid antigen test-based for SARS-CoV-2 detection in nursing homes in Valencia, Spain. J ClinVirol. 2021;143:104941.
Crossref - Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis. 2021;73(11):e3884-99.
Crossref - Rhoads D, Peaper DR, She RC, et al. College of American Pathologists (CAP) Microbiology Committee Perspective: Caution must be used in interpreting the Cycle Threshold (Ct) value. Clin Infect Dis. 2020;72(10):e685-e686.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.