ISSN: 0973-7510
E-ISSN: 2581-690X
Malaria is a global threat and a never-ending battle without appropriate identification and differentiation of the parasite species. This work compared the diagnostic methods including the thick film microscopy technique, quantitative buffy coat, and polymerase chain reaction. The inaccuracy of species determination by microscopy and the consequent treatment regime underlines the necessity to upgrade routine diagnostic methods with molecular techniques. In the study, 436 samples were collected; venous blood was processed for the quantitative buffy coat technique followed by classical Giemsa staining of thin and thick smears and nested Polymerase Chain Reaction (nPCR) for the genus-specific region of Plasmodium targeting 18S rDNA followed by species-specific identification. Of 436 samples screened for malaria, results in PCR showed 78.7% (100/127) to be P. vivax, 4.8% (6/127) as P. falciparum and 16.5% (21/127) to be mixed infection (P. vivax + P. falciparum). The prevalence of malaria was 0.29, and there was good concordance between the methods for detecting Plasmodium (Kappa:0.77). In our investigation, nested PCR and TFM exhibited a sensitivity of 97.7% and a specificity of 100% for malaria detection compared to QBC. Clinical parameters- thrombocytopenia and anemia, were compared in this study. A positive association was observed between thrombocytopenia and malaria (p<0.05), but the association between anemia and malaria infection remains unclear. Primer cross-reactions were also observed in the primer sequence of P. ovale and P. knowlesi, but sequencing confirmed it as P. vivax and the study of phylogeny paved a new way in analyzing the relatedness of the sequences.
Malaria, Anemia, Primer Cross-reaction, TFM, QBC, PCR
Malaria continues to be a major public health concern threatening 106 countries, with approximately 650,000 annual deaths.1 In 2020, an estimated 241 million cases of malaria were recorded worldwide compared to 227 million cases in 2019.2,3 About 3% of cases are reported in India.4 Malaria is very common in India, with two primary malarial parasites, Plasmodium falciparum and Plasmodium vivax, wreaking havoc.5,6 An estimated 0.7–1.6 million cases and 400-1,000 fatalities are reported annually in India. In 2019, Karnataka recorded ~3500 cases of malaria, of which ~2800 (80%) were reported from the Udupi and Dakshina Kannada districts. Mangaluru, a coastal city of Dakshina Kannada, was labeled as endemic to malaria, with P. vivax, being responsible for most cases.7 Though 90% of the reported cases were diagnosed as P. vivax, most of the mortality was seen in patients positive for P. falciparum.8,9 The emergence of severe malaria in India during 1994–1995 has significant implications for the planning of malaria control programs.
The city of Mangaluru reported an increase in malarial vectors, a consequence of increasing urbanization resulting in construction activities. P. vivax affects more than 50% of Mangaluru’s population. Initially considered a milder form of malaria, the increased morbidity with relapse has made it difficult to eradicate. In contrast to P. falciparum, P. vivax has greater prevalence and genetic variation in the district. Mortality caused by malaria goes unnoticed in India by the healthcare facilities leading to inaccurate contribution of information on the global burden.10,11 Since many patients have limited access to healthcare facilities; many cases go under-reported.12 In the outside sub-Saharan region of Africa, about 3.5%-16% of malaria-related morbidity was associated with severe vivax malaria. To date, conventional microscopic examination of Giemsa-stained blood smears is considered the gold standard diagnostic technique.13 Though this technique efficiently distinguishes various species of Plasmodium, it is limited in identifying Plasmodium ovale, which causes tertian malaria in humans and has morphological similarities with P. vivax. Diagnosis may be missed because of low parasitemia or mixed infections, and lack of adequately trained personnel, all of which make microscopy-based diagnosis challenging.14
Another method employed for malarial diagnosis is the Quantitative Buffy Coat (QBC) technique.15 This technique is mainly based on micro-centrifugation, fluorescence, and density gradient of infected erythrocytes. Training a technician to use QBC as a diagnostic tool requires a shorter duration of approximately a week compared to microscopic training which takes years for a trainee to be skilled enough to diagnose and characterize the species.16 For efficient treatment of malaria, early and accurate diagnosis is of utmost importance.17,18 Molecular technique-like PCR is commonly used for diagnosis as it is reliable, sensitive, and more specific than the thick film microscopy technique.19-21 For detecting Plasmodium, the first round of PCR using genus-specific primers followed by nested PCR for speciation is a promising technique.22,23 Though nested PCR assays can differentiate species even with low parasitemia, cross-reactivity remains a major concern in diagnosing closely related species like P. vivax, and P. ovale. and the considered fifth human malaria parasite Plasmodium knowlesi Earlier research employing the PCR diagnostic assay revealed numerous fascinating facts about the malaria parasite spread in India. The main objective of this study was to compare the various diagnostic methods for the detection of malaria and the prevalence of the species in the city of Mangaluru, an endemic zone located on the southwest coast of Karnataka, India. The study also focused on the association of malaria with some clinical parameters like thrombocytopenia and anemia, distribution of malarial cases with demographic variables like age and gender as well as distribution of Plasmodium species among positive cases identified in the study.
Study design
The study was conducted in Mangaluru, located in coastal Dakshina Kannada District, Karnataka, India, where malaria is endemic with seasonality factors influencing the vector population. P. falciparum and P. vivax infections have been observed to predominate the coastal areas. 436 blood samples were obtained during the period 2017–2019, from individuals with suspected malarial fever from a major hospital in the region with more than ten thousand patients visiting the outpatient department on a monthly average. Patients of the age group more than 2 years, with fever and any two of the following conditions, chills, sweating, jaundice, splenomegaly, convulsions, coma, shock, and pulmonary edema, were included in the study as per the criteria mentioned in the National Vector Borne Disease Control Programme of India (www.nvbdcp.gov.in ).
Ethics approval
This study was approved by the Institutional Scientific and Central Ethics committee (vide; NU/CEC/ICMR-05/2015, NU/CEC/2018/0183) of Nitte Deemed to be University, Mangaluru and permitted by District Health Officer, Mangaluru (vide; No: AV (2) 283/2018-19). The malaria microscopy training for the study was imparted by the Government Wenlock District Hospital, a recognized center under the National Vector Borne Disease Control Programme of the District Mangaluru (Dakshina Kannada), India (vide; No/DVBDCP/108/2018-19).
Blood Collection, Quantitative Buffy Coat (QBC) analysis, and Thick Film Microscopy (TFM)
Venous blood (2mL) was collected in a sterile heparinized tube from the febrile patients and screened for malaria parasite by QBC and microscopic examination of the stained smear. Blood parasite detection by QBC15,24 is based on centrifugal stratification of blood elements and was used for blood parasite detection. Additionally, 2–3 drops of blood were used for thin and thick smear preparation on microscopic slides, followed by Giemsa staining and microscopic examinations by a lab technician skilled in malarial microscopy.25 The thick film counted parasites among 200 leukocytes (WBCs). The density of the parasites was obtained by estimating a cumulative WBC count of 8000/mL and 4.5 million RBC/mL and examining at least 200 fields before recording the result as negative.26
Thrombocytopenia and Anemia
Platelet count <150,000 platelets/µL was classified as thrombocytopenia.27 Anemia was considered to be so when the hemoglobin was < 11 g/dL. The samples were categorized as platelet counts less than <150,000 platelets/µL and more than 150,000 platelets/µL. Likewise, Anemia is defined by the World Health Organization (WHO) as hemoglobin (Hb) levels of 12.0 g/dL in women, and 13.0 g/dL in men,28 less than this value is considered anemic and more than being non-anemic. These data were analyzed to compare thrombocytopenia and anemia association with malaria.
Polymerase chain reaction and Molecular analysis
For molecular studies, ~200 µL of heparinized whole blood sample was used for the total DNA extraction using DNeasy Blood and Tissue Kit (QIAGEN). PCR was performed using the previously published primers.22, 29 For the genus-specific detection, the following primers were used: rPLU1/rPLU5 for the primary (first step) and rPLU3/rPLU4 for secondary (nested) amplification. Each 30 µL reaction mixture contained 2 µL (100 ng/µL) of DNA template, 0.8 µL (8 8 pM) of each primer, 3.3 microliters 10x buffer, 0.8 µL (110 mM) of deoxynucleoside triphosphate, and 0.3µL (1.5 units) of Taq DNA polymerase. Primary amplification comprised subjecting the reaction mix to an initial denaturation at 94°C for 3 minutes followed by 40 cycles that included denaturation at 94°C for 60 seconds, annealing at 54°C for 30 seconds, extension at 72°C for 90 seconds, and final delay at 72°C for 10 minutes. The samples that showed amplification using rPLU1/rPLU5 primers were tested for species-specific primers rVIV1/rVIV2 for P. vivax, rFAL1/rFAL2 for P. falciparum, rMAL1/rMAL2 for P. malariae, pmk8/pmk9 and Pk1140/Pk1150 for P. knowlesi 29 and rOVA1/rOVA2 and rOVA 3/rOVA4 for P. ovale.30,31 Two microliters (100ng/µL) of the primary amplification product served as the template for each 30-µL nested PCR reaction. The concentrations of the nested reaction primers and other constituents were the same as those for primary amplification. Nested reaction conditions were also identical to primary amplification except that the annealing temperature for genus-specific primers rPLU3 and rPLU4 was 52°C and varied for species-specific primers, with 60°C for rFAL1/rFAL2, 53.5°C for rMAL1/rMAL2, 55°C for rVIV1/rVIV2, 50°C for rOVA3/rOVA4, 48°C for pmk8/pmk9. The PCR products of the nested reaction were analyzed by gel electrophoresis, stained with ethidium bromide, and visualized under UV-Transilluminator (Biorad, Germany).
Sequence analysis
The PCR products were outsourced for sequencing after purification with a QIAquick PCR purification kit (QIAGEN, Germany), and the sequences were analyzed using MEGA X and Clustal W. The sequences were aligned with the reference 18S rDNA gene of the 3D7 isolate of P. falciparum (GenBank accession no. AL844501) and SAL-1 strain of P. vivax (GenBank accession no. U03079.1) using the BLAST search.
Statistical analysis
Descriptive figures were expressed as medians and frequencies. Samples were categorized based on age, gender, the status of anemia, and thrombocytopenia with the occurrence of malaria infection. Each of the three diagnostic techniques’ sensitivity, specificity, and predictive value were computed by comparing them to a composite reference21 generated from the three methods. Sensitivity, specificity, statistical positives and negatives, accuracy, and percentage of agreement (kappa value) were calculated with confidence intervals.32 Plasmodium genus/species positive in all the methods were considered as True Positive (TP), and negative by all methods were True Negative (TN), samples wrongly detected as positive in a particular method were False Positive (FP), and those detected as negative was considered False Negative (FN). Calculations of sensitivity, specificity, and predictive values of diagnostic methods were performed using the following formulae: sensitivity = (TP / (TP + FN)) × 100, specificity = (TN/ (TN + FP)) × 100, positive predictive value = (TP / (TP + FP)) × 100, negative predictive value = (TN/ (TN + FN)) × 100.33 Chi-square tests were performed to find an association between the categorical variable (gender, status of anemia, and thrombocytopenia) with the occurrence of malaria independently. Statistical analysis was performed using SPSS software ver. 29.0 (IBM, Armonk, NY, USA) on the data, p < 0.05 was considered significant.
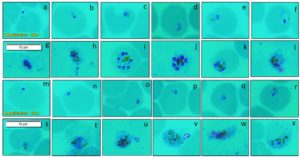
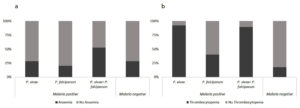
Microscopic examination showed Plasmodium in different life-cycle stages of the parasite. Based on the morphologically distinguishable features of microscopic observation as explained in WHO basic malaria microscopy guidelines,26 the samples were further characterized into P. vivax, P. falciparum, and mixed infection (P. vivax + P. falciparum). Figure 1 depicts the Giemsa-stained erythrocytic developmental stages of malarial parasites for two species, P. vivax, and P. falciparum (Figure 1). Among the confirmed malaria cases (127/436) all the patients had a fever, chills /sweating as primary symptoms and other presenting. symptoms like jaundice (5%), and splenomegaly (3%), none of the other patients reported coma, shock, or pulmonary edema. One patient presented with cerebral malaria without symptoms of convulsions. Of the 436 suspected malaria, samples collected, 290 (66.6%) were males, and 148 (33.4%) were females. The positive cases were categorized based on age and gender (Supplementary file 1). It was found that the male population was significantly associated with positive malaria cases (p<0.05), however, the age factor was not associated with positive cases. Thrombocytopenia, and anemia and their relation with the status of malaria was analyzed. Of 91 infected patients positive for P. vivax, 84 (92%) were thrombocytopenic (Figure 2a), and 26 (28.5%) were anemic
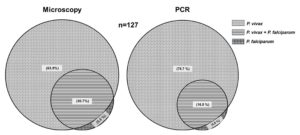
(Figure 2b). Similarly, among 5 P. falciparum-infected patients, 2(40%) were thrombocytopenic, and only 1(20%) was anemic. In cases of mixed infection by P. vivax and P. falciparum of 19, 17 (89%) were thrombocytopenic, and 10 (52%) were anemic, we lacked patient information of anemia and thrombocytopenia for 12 samples that were positive for malaria. In cases of febrile illness (n=309), we lacked patient details 80 samples in anemia and 55 samples in thrombocytopenia. A positive association was observed between thrombocytopenia (p<0.05) and malaria, but the association between anemia and malaria infection was non-significant (p>0.05). TFM results indicated that 81 samples (63.8%) were positive for P. vivax, 39 (30.7%) were positive for mixed species (P. vivax + P. falciparum), and 7 (5.5 %) were positive for P. falciparum of 127 positives and in QBC findings showed that 69 samples (54 %) were positive for P. vivax, 52 (40.5 %) were positive for mixed species (P. vivax + P. falciparum), and 7 (5.5%) of the 128 positives were positive for P. falciparum.
Figure 1. Microscopic image showing Giemsa’s stained erythrocytic developmental stages of malarial parasites. Image a-l: P. vivax and image m-x: P. falciparum Trophozoite stages to Gametocyte stages
Figure 2. Bar diagram showing the population affected. a. Distribution of thrombocytopenia present or absent among the species b. Distribution of anaemia present or absent among the species
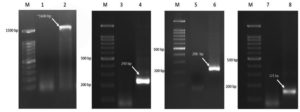
PCR was performed for all 436 samples using primers mentioned in the methodology. The genus-specific primers targeting 18S rDNA were expected to generate an amplicon of ~1600 bp, which was further used for nested PCR to confirm the genus and species. A 240 bp amplicon was obtained in the positive samples confirming the Plasmodium genus. The primers used for P. vivax generated a product of 121 bp and primers targeting P. falciparum generated a product of 206 bp (Figure 3). In the case of P. ovale and P. knowlesi, the respective primers generated amplicons of 456 bp and 153 bp, respectively. Results of PCR revealed 100 samples (78.7%) positive for P. vivax, 21 (16.5%) showing mixed species (P. vivax + P. falciparum), and 6 (4.8%) positive for P. falciparum (Figure 4).
Figure 3. Representative gel image of PCR characterization Plasmodium species. Lanes M:100 bp DNA ladder, Lane 1,3,5&7: Non-Template Control (NTC); Lanes 2&4: Plasmodium genus 18S rDNA primary and secondary amplification respectively; Lanes 6&8: Species confirmation band of P. falciparum and P. vivax respectively
Figure 4. Venn diagram showing the distribution of Plasmodium species. In microscopy (TFM) out of 127 positive malaria cases, 81 samples (63.8%) were positive for P. vivax, 39 (30.7%) were positive for mixed species (P. vivax + P. falciparum), and 7 (5.5 %) were positive for P. falciparum, whereas in PCR showed 100 samples (78.7%) positive for P. vivax, 21 (16.5%) showing mixed species (P. vivax + P. falciparum), and 6 (4.8%) positive for P. falciparum
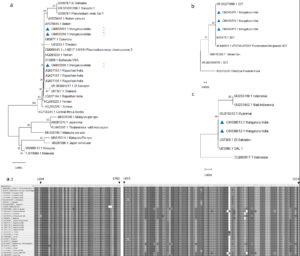
The prevalence of malaria was 0.29 (127/436) considering positives (true positive) by all the methods, with 97.67% sensitivity and 100% specificity for both PCR and TFM, whereas 98.45% sensitivity and 99.67% specificity were obtained for the QBC method when each diagnostic method was compared to the composite reference (Supplementary file 2). All the diagnostic methods (TFM, QBC, and PCR) confirmed 127 samples as positive for malaria. The number of P. vivax samples detected by PCR and TFM was 70. The number of mixed samples detected by both PCR and TFM was 9. However, 28 samples that were detected as a mixed infection through TFM were found to be positive only for P. vivax. A good concordance between both methods for detecting Plasmodium at the genus level (Kappa:0.77) with a 95% confidence interval is observed. To confirm the 18S rDNA PCR amplicons, twelve PCR products that included two P. vivax, five P. falciparum, three P. ovale, and two P. knowlesi were sequenced. The sequences obtained were aligned with the reference sequence using suitable computational tools mentioned in the methods. The sequencing results provided interesting information. Three samples reported as positive for P. ovale and two for P. knowlesi turned out to be P. vivax. The interesting sequences thus generated with a cross-reactivity were also deposited in the GenBank and assigned accession numbers. Phylogenetic analysis of partial sequences was constructed with the maximum likelihood estimation model using the MEGA X program (Figure 5). All sequences of P. vivax deposited included GenBank accession no. OM033487 & OM033488, OM033596 – OM033598, & OM038613, and P. falciparum GenBank accession no. OM057670 & OM057671, OM049473- OM049475.
The phylogenetic tree is represented in Figure 5a from sequences amplified using P. ovale 18S primers rOVA 3/rOVA4, which amplified regions of P. vivax 18S rDNA showing cross-reactivity of primer. Two sequences (accession number: OM033597, OM033598) showed similarity to the reference Belem strain and the Sal 1 from Brazil, with the region covering 406 bp. These two sequences had nucleotide A at the 1519 position, similar to the reference sequences. Another sequence (accession number: OM033596) amplified using the same primer showed similarity to sequences from Rajasthan, India. A1519T substitution was unique in the accession number mentioned above from India similar to Sal 1. P. falciparum sequences are amplified using nested primers rPLU1/rFAL2 (Figure 5b) and rFAL1/rFAL2 yielded 829 bp and 205 bp, respectively. The amplified region was highly similar to the 18S rDNA gene of the 3D7 reference sequence. P. vivax-specific primers (rVIV1/rVIV2), which amplified 121 bp and the submitted sequences (accession number: OM033487, OM033488) were highly similar to the reference sequence (Figure 5c).
Figure 5. Phylogenetic analysis of 18S rDNA gene of P. vivax and P. falciparum partial sequences. The phylogeny tree was constructed with the Neighbor-joining method using the MEGA X program
The set of primers that also exhibited cross-reactivity upon using primers for P. knowlesi (pmk8/pmk9) amplified 153 bp from P. vivax (accession number: OM038612, OM038613), which was similar to the P. vivax Salvador I (XR_003001211) reference sequence.
The current study was conducted in a malaria-endemic zone using three different methods for malaria diagnosis. Comparisons made included clinical associations based on laboratory parameters, and diagnosis using TFM with or without QBC. Malaria misdiagnosis is common when there is low parasitemia, and when antimalarial drugs are not specific to the species causing malaria.34-37 The misdiagnosis leads to an inappropriate prescription of high-cost medications with the patient exposed to potentially toxic drugs.38-40 The success of TFM in malaria diagnosis depends upon a high level of personnel competency for accuracy.41 An experienced and well-trained technician is the key to TFM and is considered the gold standard in diagnosing malaria. Recently, the use of molecular tests such as PCR is highly acclaimed and therefore popular for the sensitive identification and differentiation of Plasmodium species simultaneously.19 However, the success of this method depends on the specificity of the primers. In this study, 127 of 436 samples were confirmed as malaria by the three methods, demonstrating the usefulness of TFM, QBC as well as PCR. However, it is crucial to differentiate the species causing the type of malaria when reporting positive in patients for accurate treatment and to use the right anti-malarial drugs, especially in endemic areas.13,42 The inaccuracy of species determination by thick film microscopy has underlined the necessity for sufficient training and regular skill monitoring for microbiologists involved in malaria diagnosis, as well as an upgrade to the routine use of PCR-based molecular techniques. In this study, 10 cases diagnosed as P. vivax by TFM were later confirmed by PCR as a mixed infection due to P. vivax and P. falciparum. In contrast, 28 cases diagnosed as a mixed infection turned out to be mono-infection by P. vivax. One case confirmed as P. falciparum by TFM was confirmed as P. vivax by PCR. The importance of early detection and treatment is paramount to avoiding complications in malaria. It has been observed that cerebral malaria patients affected by P. falciparum who do not receive prompt treatment end up with unnecessary complications leading to death. Nested PCR has higher sensitivity and specificity than TFM. This is endorsed in our study in which nested PCR for malaria detection exhibited a sensitivity of 97.67 percent and specificity of 100 percent compared to the microscopic examination of Giemsa-stained smears, especially for species determination for accurate treatment.
Earlier studies using PCR for malaria detection23,30,31,43,44 have demonstrated the superiority of the technique as they observed cross-reactivity between some species that occurred in differentiating P. knowlesi and P. ovale from P. vivax using primers pmk8 and pmk9 as well as rOVA3 and 4. We also detected eight positive samples for P. vivax by PCR and generated amplicons with primer pairs pmk8 and pmk9. Likewise, 35 samples that were positive for P. vivax generated amplicons suggesting P. ovale using the primer pair rOVA3 and rOVA4. Since the gene regions used earlier were known to result in cross-creativity, for further confirmation, we performed PCR with new sets of primers released by the same authors, after encountering cross-reactivity, and the primer that was suggested as promising alternatives.23,30,43 This region was believed to help identify P. knowlesi and P. ovale using semi-nested PCR amplification of a fragment of DNA that differed in the target region from that of P. vivax. Our results showed the samples as positive for vivax only, ruling out the presence of P. ovale or P. knowlesi in the 127 positive samples. The sequencing results further confirmed the samples as positive for P. vivax. The results highlighted the genomic variation in the primer binding region of P. vivax which could be the primary reason for cross-reactivity, some of the primers we used exhibited cross-reactivity using primers for P. knowlesi (pmk8/pmk9), and primers for P. ovale (rOVA 3/rOVA4) indicating the need for a review of existing primers for species identification. The phylogenetic tree constructed using the sequence, revealed the diversity in the circulation of malaria parasites in this endemic region, which could be due to the migration of workers in this geographical area, especially among people involved mainly in urban development. It could also be attributed to the P. vivax and P. falciparum mixed infections seen predominantly in the area.
We also examined the association of malaria with clinical parameters like thrombocytopenia and anemia, the distribution of Plasmodium cases with demographic variables like age and gender as well as the distribution of Plasmodium species in positive cases identified in the study. An increased infection rate was seen in the active age group of 20-29. A similar trend was also observed among the female population. However, no correlation could be drawn with the existing data. The dominance of the P. vivax species in Mangaluru followed by a mixed infection involving P. vivax and P. falciparum was observed. This trend was previously observed in Mangaluru by Dayanand et al.45 from January 2013 through December 2016. The least prevalent species reported in this study were P. falciparum. An observation that thrombocytopenia or anemia is a hematological complication associated with malaria has been reported.46-47 In our study, the majority of the positive malaria patients showed thrombocytopenia. ~84% of the negative samples showed no signs of thrombocytopenia, thereby highlighting its use as a predictor or indicator of malaria. However, this pattern was not observed in the case of anemia, where about ~75% of the non-malarial febrile patients showed anemic conditions. In comparison, only 32% of the patients who tested positive for malarial parasites had anemia. Though this association does not throw light on the use of thrombocytopenia as a marker, it remains to be seen whether these hematological conditions are a cause or consequence of malaria.
The inaccuracy of species determination by thick film microscopy has underlined the necessity to upgrade routine diagnostic methods with complementary molecular techniques. PCR-based strategies coupled with genomic sequencing will be a viable method for the detection and differentiation of species in malarial cases, especially for early and accurate diagnosis when the parasitemia is low for directing appropriate treatment. The Association of clinical parameters with malaria can be explored to understand the pathogenesis and complications that arise due to severity and in chronic cases.
Additional file: Additional Table S1. Additional Figure S1.
ACKNOWLEDGMENTS
The authors would like to thank Nitte University Centre for Science Education & Research and Nitte (Deemed to be University) for providing a Research environment and facility for carrying out the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
IK, PR and RA conceptualized and designed the study. AR performed literature review. AR and TS collected the data. AR performed analyses. AR and AK wrote the original draft. AR, AK, and IK edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was supported by Nitte (Deemed to be University), Nitte University Research grant (NUFR2/2018/10/26).
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Scientific and Central Ethics committee (vide NU/CEC/ICMR-05/2015, NU/CEC/2018/0183) of Nitte Deemed to be University, Mangaluru and also permitted by District Health Officer, Mangaluru (No: AV (2) 283/2018-19).
- World Health Organization. World malaria report 2021. https://www.who.int/publications/i/item/9789240040496, Accessed 6 December 2021.
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64(1-2 Suppl):97-106.
Crossref - Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4(6):327-336.
Crossref - Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006;22(8):353-358.
Crossref - Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77(6 Suppl):79-87.
Crossref - Murray CJ, Ortblad KF, Guinovart C, et al. Global, regional, and national incidence and mortality for HIV, tuberculosis, and malaria during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013 Lancet. 2014;384(9947):1005-1070.
Crossref - Das A, Anvikar AR, Cator LJ, et al. Malaria in India: the center for the study of complex malaria in India. Acta Trop. 2012;121(3):267-273.
Crossref - Joshi H, Prajapati SK, Verma A, Kang’a S, Carlton JM. Plasmodium vivax in India. Trends Parasitol. 2008;24(5):228-235.
Crossref - Saravu K, Kumar R, Ashok H, et al. Therapeutic Assessment of Chloroquine-Primaquine Combined Regimen in Adult Cohort of Plasmodium vivax Malaria from Primary Care Centres in Southwestern India. PLoS One. 2016;11(6):e0157666.
Crossref - Dhingra N, Jha P, Sharma VP, et al. Adult and child malaria mortality in India: a nationally representative mortality survey. Lancet. 2010;376(9754):1768-1774.
Crossref - Hay SI, Gething PW, Snow RW. India’s invisible malaria burden. Lancet. 2010;376(9754):1716-1717.
Crossref - Gupta P, Sharma R, Chandra J, Singh V. Severe malaria due to Plasmodium vivax: case report. Curr Pediatr Res. 2016.
- Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8.
Crossref - Ohrt C, Purnomo, Sutamihardja MA, Tang D, Kain KC. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J Infect Dis. 2002;186(4):540-546.
Crossref - Clendennen TE 3rd, Long GW, Baird JK. QBC and Giemsa-stained thick blood films: diagnostic performance of laboratory technologists. Trans R Soc Trop Med Hyg. 1995;89(2):183-184.
Crossref - Kochareka M, Sarkar S, Dasgupta D, Aigal U. A preliminary comparative report of quantitative buffy coat and modified quantitative buffy coat with peripheral blood smear in malaria diagnosis. Pathog Glob Health. 2012;106(6):335-339.
Crossref - Hanscheid T. Current strategies to avoid misdiagnosis of malaria. Clin Microbiol Infect. 2003;9(6):497-504.
Crossref - Cuadros J, Martin-Rabadan P, Merino FJ, Delgado-Irribarren A, Garcia-Bujalance S, Rubio JM. Malaria diagnosis by NOW ICT and expert microscopy in comparison with multiplex polymerase chain reaction in febrile returned travellers. Eur J Clin Microbiol Infect Dis. 2007;26(9):671-673.
Crossref - Snounou G, Viriyakosol S, Zhu XP, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61(2):315-320.
Crossref - Pakalapati D, Garg S, Middha S, et al. Comparative evaluation of microscopy, OptiMAL(®) and 18S rRNA gene based multiplex PCR for detection of Plasmodium falciparum & Plasmodium vivax from field isolates of Bikaner, India. Asian Pac J Trop Med. 2013;6(5):346-351.
Crossref - Berzosa P, de Lucio A, Romay-Barja M, et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar J. 2018;17(1):333.
Crossref - Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60(4):687-692.
Crossref - Singh B, Kim Sung L, Matusop A, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017-1024.
Crossref - Ahmed NH, Samantaray JC. Quantitative buffy coat analysis-an effective tool for diagnosing blood parasites. J Clin Diagn Res. 2014;8(4):DH01.
Crossref - Mfuh KO, Achonduh-Atijegbe OA, Bekindaka ON, et al. A comparison of thick-film microscopy, rapid diagnostic test, and polymerase chain reaction for accurate diagnosis of Plasmodium falciparum malaria. Malar J. 2019;18(1):73. Published 2019 Mar 12.
Crossref - World Health Organization, Center for Disease Control. Basic malaria microscopy: tutor’s guide. World Health Organization; 2010.
- Martinez-Salazar EL, Tobon-Castano A. Platelet profile is associated with clinical complications in patients with vivax and falciparum malaria in Colombia. Rev Soc Bras Med Trop. 2014;47(3):341-349.
Crossref - Cappellini MD, Motta I. Anemia in Clinical Practice-Definition and Classification: Does Hemoglobin Change With Aging? Semin Hematol. 2015;52(4):261-269.
Crossref - Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26(2):165-184.
Crossref - Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol. 2009;47(12):4173-4175.
Crossref - Wang B, Han SS, Cho C, et al. Comparison of microscopy, nested-PCR, and Real-Time-PCR assays using high-throughput screening of pooled samples for diagnosis of malaria in asymptomatic carriers from areas of endemicity in Myanmar. J Clin Microbiol. 2014;52(6):1838-1845.
Crossref - Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977;33(2):363-374.
Crossref - Ojurongbe O, Adegbosin OO, Taiwo SS, et al. Assessment of Clinical Diagnosis, Microscopy, Rapid Diagnostic Tests, and Polymerase Chain Reaction in the Diagnosis of Plasmodium falciparum in Nigeria. Malar Res Treat. 2013;308069.
Crossref - Ghai RR, Thurber MI, El Bakry A, Chapman CA, Goldberg TL. Multi-method assessment of patients with febrile illness reveals over-diagnosis of malaria in rural Uganda. Malar J. 2016;15(1):460.
Crossref - Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg. 2007;77(6 Suppl):119-127.
Crossref - Wilson ML. Laboratory diagnosis of malaria: conventional and rapid diagnostic methods. Arch Pathol Lab Med. 2013;137(6):805-811.
Crossref - Baird JK, Valecha N, Duparc S, White NJ, Price RN. Diagnosis and Treatment of Plasmodium vivax Malaria. Am J Trop Med Hyg. 2016;95(6 Suppl):35-51.
Crossref - Amexo M, Tolhurst R, Barnish G, Bates I. Malaria misdiagnosis: effects on the poor and vulnerable. Lancet. 2004;364(9448):1896-1898.
Crossref - Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229-241.
Crossref - Ghai RR, Thurber MI, El Bakry A, Chapman CA, Goldberg TL. Multi-method assessment of patients with febrile illness reveals over-diagnosis of malaria in rural Uganda. Malar J. 2016;15(1):460.
Crossref - Ashraf S, Kao A, Hugo C, et al. Developing standards for malaria microscopy: external competency assessment for malaria microscopists in the Asia-Pacific. Malar J. 2012;11:352.
Crossref - Kaewkamnerd S, Uthaipibull C, Intarapanich A, Pannarut M, Chaotheing S, Tongsima S. An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinformatics. 2012;13(Suppl 17):S18.
Crossref - Ndao M, Bandyayera E, Kokoskin E, Gyorkos TW, MacLean JD, Ward BJ. Comparison of blood smear, antigen detection, and nested-PCR methods for screening refugees from regions where malaria is endemic after a malaria outbreak in Quebec, Canada. J Clin Microbiol. 2004;42(6):2694-2700.
Crossref - Anthony C, Mahmud R, Lau YL, Syedomar SF, Sri La Sri Ponnampalavanar S. Comparison of two nested PCR methods for the detection of human malaria. Trop Biomed. 2013;30(3):459-466.
- Dayanand KK, Kishore P, Chandrashekar V, et al. Malaria Severity in Mangaluru City in the Southwestern Coastal Region of India. Am J Trop Med Hyg. 2019;100(2):275-279.
Crossref - Khan SJ, Abbass Y, Marwat MA. Thrombocytopenia as an indicator of malaria in adult population. Malar Res Treat. 2012;2012:405981.
Crossref - Akhtar MN, Jamil S, Amjad SI, Butt AR, Farooq M. Association of malaria with thrombocytopenia. Ann King Edw Med Univ. 2005;11(4).
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.