ISSN: 0973-7510
E-ISSN: 2581-690X
Bacterial lipases are the most important collection of biocatalysts used for a variety of biotechnological applications. In the current study the morphological, biochemical and Molecular characteristics to the extracellular lipase producing bacteria were identified. The bacterial isolates were selected as lipase producing bacteria using Rhodamine-B agar plate. the production of Lipase from the organism were determined with varying incubation temperature (20 to 50°C) and range of incubation time (16-120) hrs. 16 bacterial isolates were identified as Staphylococcus hemolytic. One isolate “St1” was identified By comparing the observed DNA sequences of these local samples with the retrieved DNA sequences (GenBank acc. MG595372.1) as showed high lipase activity according to experimental conditions about 140 U/ml. The current study showed that enzymatic activity of crude lipase extract was maximum in 72 hrs incubation period and 37°C.
Lipase, Staphylococcus hemolytic, Genetic Identification, Lipase extraction, Enzyme activity.
Lipases were occupied a special situation as biotechnologically valuable enzymes and display vital roles in various industries1. Lipases occur widely in nature and only microbial lipases are substantial. Microbial lipases are often more useful than enzymes derived from plants or animals because of the great variety of catalytic activities available, the high yields possible, ease of genetic manipulation, regular supply due to absence of seasonal fluctuations and rapid growth of microorganisms on inexpensive media2. Lipase is an enzyme with considerable commercial and industrial importance. Lipase (triacylglycerol acyl hydrolases (E.C.3.1.1.3) belongs to the class of hydrolases which catalyze the hydrolysis of triglycerides to glycerol and free fatty acids over an oil-water interface3. Lipases catalyze the reverse reactions in the synthesis of esters (triacylglycerols) consist of glycerol and fatty acids of long-chain in non-aqueous circumstances4. The activities of All lipolytic enzymes are depended on the pattern of α/β- hydrolase fold, However, the lipases from microbial origin specially show a distinctive folding design of α/β- hydrolase fold5,6. The greater part of industrial enzymes are produced from microbial source, they are more stable and constitute a superior diversity of catalytic actions7.
Staphylococci spp. release extracellular lipases in a fermentation medium were encompassed remedying and degrading enzymes in the environments8. The present study aimed to determine the production of lipase from bacterial isolates by investigation the produced lipase from clinical specimens and determination its activity in these isolates.
Isolation of lipase producing isolates
The clinical isolates included in the present study were collected within a one-month period. The isolates were obtain from unite of clinical microbiology in the hospital training in the governorate. These isolates were taken from primary isolation plates set up on solid media such as blood agar, MacConkey agar and mannitol salt agar. The bacterial colonies which observed on nutrient agar medium were isolated to identification and selected for screening lipase activity9.
Identification of bacterial isolates
The morphological, biochemical and genetic features of isolates were identified according to Bergey’s manual of determinative bacteriology.
DNA Sequencing of PCR amplicons
The bacterial isolate which shows maximal lipase activity on ROA plates assay is considered as positive colonies for lipase enzyme production, the isolates were identified according to molecular genetic study by using of 16SrRNA gene sequencing.
The resolved PCR amplicons were sequenced from both termini of forward and reverse based to the sequencing company instruction manuals (Macrogen Inc. Geumchen, Seoul, South Korea). By comparing the observed DNA sequences of local bacterial samples with the retrieved neighboring DNA sequences of the NCBI Blastn engine.
The results of the PCR products sequencing of the isolate was analyzed as long as with the respective sequences in the reference database. The observed variations in each sequenced sample were numbered in PCR amplicons as well as in its corresponding position within the referring genome.
A specific comprehensive bacterial tree was constructed in this study according to the protocol described by10. The observed bacterial variants were compared by reference sequences neighbor homologous by means of NCBI-BLASTn server11. Then, the results of BLAST of the observed variations were linked and aligned together using a Clustal Omega based tools. A full inclusive tree, including the observed variant, was imagined according to (http://tree.bio.ed.ac.uk/software/figtree/)12.
Lipase producing bacteria investigation
The Rhodamine B- olive oil-agar medium (ROA) was used to select the lipase producing isolates. The medium was prepared according to13. The isolates were identified after 48h of incubation at 37°C and exposed to U.V. light at 350 nm14. The isolates were culture overnight in liquid medium was prepared according to15.
The standard liquid medium for measurement the lipase activity according16 use for culturing lipase producing bacteria. the broth media were inoculated of 5% overnight culture media of all the lipase producing isolates was prepared according to17, the medium consist of (g/L): yeast extract, 2.5; pepton, 5.5; NaCl, 10 and 1% olive oil, autoclaving at 121°C for 15 min. The media incubated at 150 rpm, 37°C and pH value 6 18.
Lipase crude extraction
The crude lipase was extracted according to16.
Lipase assay
The titrimeteric method was used to measure the activity of lipase based on18. using olive oil as substrate.
Effecting the incubation period and incubation temperature in lipase production by isolate Staphylococcus hemolytic
A selection of isolate Staphylococcus hemolytic was cultivated in fermentation medium for five day. varying from 20°C to 50°C of incubation temperature were selected to determine the optimum temperature for lipase production, while other limits keeping at same19.
The laboratory and genetic identification
The technique of Enrichment culture enable the isolation strains of bacteria from different source. The results in the current study appear that the supposed isolate was Staphylococcus hemolytic according to its’ identifying as Gram positive, cocci in clusters and tests made for catalase and coagulase production, allowing the coagulase-positive staphylococci to be identified quickly20.
Within this locus, The sample was included in the present study that had shown to amplify 1429 bp genetic sequences in Staphylococcus haemolyticus genome. The sequencing reactions indicated that the exact identity after performing NCBI blastn for these PCR amplicons “https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch”. Concerning the supposed ribosomal 1429 bp amplicons, NCBI BLASTn engine shown about 100% sequences similarities between the sequenced samples and the intended reference target sequences. By comparing the observed DNA sequences of these local samples with the retrieved DNA sequences (GenBank acc. MG595372.1), the approximate positions and other details of the retrieved PCR fragments were identified (Fig. 1).
Fig. 1. The exact position of the retrieved 1429 bp amplicon that partially covered a portion of 16S rRNA locus of Staphylococcus haemolyticus genomic DNA sequences (GenBank acc no. MG595372.1).
After positioning the 1429 bp amplicons’ sequences, the details of its sequences were highlighted, in terms of the positioning of both forward and reverse primers of the 1429 bp amplified amplicon (Table 1).
Table (1):
The position and length of the 1429 bp PCR amplicons used to amplify a portion of 16S rRNA locus of Staphylococcus haemolyticus genomic DNA sequences (GenBank acc no. MG595372.1).
Amplicon |
Referring locus sequences (5′ – 3′) |
length |
|---|---|---|
16SrRNA sequences |
ATACATGCAGTCGAGCGAACAGATAAGGAGCTTGCTCCTTTGACGTTA
|
1429 bp |
The alignment results of the 1429 bp samples revealed the absence of any detected polymorphism in this position in the investigated sample in comparison with the referring reference DNA sequences (Fig. 2).
Fig. 2. DNA sequences alignment of the local sample of 16S rRNA with their corresponding reference sequences of the 1429 bp amplicons within Staphylococcus haemolyticus genomic DNA.
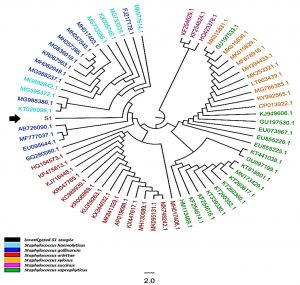
A comprehensive phylogenetic tree was constructed, (65) sequences were comprised the total number of the aligned nucleic acid sequences, irrespective of the investigated local bacterial variant. The total number of the included species within this tree was made of six species within Staphylococcus genus, namely S. haemolyticus, S. gallinarium, S. arlettae, S. xylosus, S. succinus, and S. saprophyticus. Though the current constructed comprehensive tree indicated the presence of several species one, one genus was detected allover this comprehensive tree (Figure 3). However, it was revealed that the investigated St1 was positioned in a unique position beside KT026096.1 and AB726090.1 accession number which they belonged to S. haemolyticus and S. gallinarium respectively. This observation gave another confirmed ideation with regard to the accurate identity of S. hamelotyicus. However, the unique position occupied by the observed S1 between S. haemolyticus and S. gallinarium has indicated a particular positioning of the St1 in terms of its phylogenetic association with the other related species within the Staphylococcus genus. However, this multi-species positioning did not mean the inability of this 16S- based ribosomal amplicon in the detection of the mainly targeted S. haemolyticus species. Moreover, the currently utilized ribosomal primers may not reflect high specificity of the targeted site as five other non-targeted organisms were revealed in the immediate vicinity to the investigated St1 isolate. This is due to the fact that St1 has characterized with unique nucleic acid sequences that made it localize in this phylogenetic position beside several related nucleic acid sequences that are belonging to other organisms of the same genus.
Fig. 3. The comprehensive phylogenetic tree of the 16S rRNA genetic fragments for Staphylococcus haemolyticus local isolates. The black color and arrows refer to the sequenced query local samples, while other colors refer to other referring NCBI Staphylococcus deposited referring species.
Investigation, screening of lipase producing bacteria
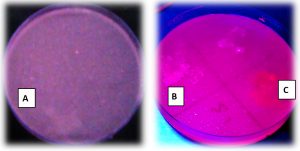
The investigation of lipase activity to the screened isolates were showed in table 2 and Fig. 4, It appears that the colonies with lipolytic activity showed orange florescence halos after growth on Rhodamine -B agar plates test for 48hr of incubation. The results showed that one bacterial isolate St1 from Sixty isolates were characterized by this activity during the present study.
Table (2):
Lipase activity on ROA plate test for isolates Staphylococcus haemolyticus.
Symbol of isolate |
Result of ROA test |
|---|---|
St1 St2 St3 St4 St5 St6 St7 St8 St9 St10 St11 St12 St13 St14 St15 St16 |
+ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ |
The hospital environment is one of different sources for a microbial population. As shown in Fig. 4 the colonies of S. haemolyticus (St1) showed positive on Rhodamine -B agar plates test. The colonies of lipase producing bacteria were showed halos of orange fluorescent during 48hrs of incubation at 37°C, This appearance may be due to the hydrolysis of substrate, or due to the construction of conjugate of a Rhodamine -B long chain fatty acid. The results in the present study agree with study of21,13,22-26. Qualitative screening of Pseudomonas sp LSK25 lipase showed positive evidence of lipase production. The strain was able to produce orange fluorescent halos (Rhodamine B) within 48 h incubation at 4°C 27.
Determination of lipase activity
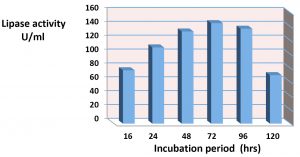
The results of effect incubation time in the lipase construction is appear in the Fig. 5. As it shown in this figure the maximum production of crude lipase extract for the isolate Staphylococcus haemolyticus in 72hrs with fixing other experimental conditions was about 140U/ml. The extracellular lipase production of Staphylococcus spp. were affected by number of factors. The present study agree with28, who suggest that 48hrs of incubation was the optimum period for lipase production. The study of29,30 showed that the maximum lipase production was found at 48hrs. of growth for Bacillus spp. The study of30 showed that the maximum production of lipase being highest than 24U/ml after 48hrs.of incubation. According to19, The maximum lipase activity by S. aureus of the culture medium was seen after 45 hours at 35°C. While the study of27 reported that The highest lipase activity was achieved over 36 h at 10°C in production medium at pH 7.0.
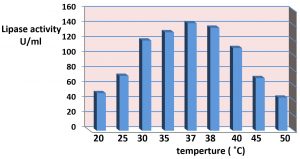
The results of effect of the incubation temperature is appear in the Fig. 6. It was showed that 37°C the optimum temperature for lipase production. The synthesis of enzyme is control by the incubation temperature of fermentation medium at mRNA transcription level and probably translation levels of enzymes thus increases the stability of enzymes and enzyme production. However the optimal temperature for lipase production agrees with the growth temperature of the microbes23. also the present study suggest that the production of lipase at high temperature due to that temperature stimuli the secretion of extracellular enzymes by shifting the physical characteristics of the cell membrane23. The study of31 reported that the effect of temperature on enzyme activity is attributed to its effect on stability, rate of the reaction, substrate solubility as well as direct influence on esterification reaction. The results in the present study agree with9,32 it reported the optimum incubation temperature for lipase production at 37°C of Pseudomonas gessardii and certain species of Pseudomonas. While the study of33-35 were reported the incubation temperature at 25°C was gave the maximum lipase production for Pseudomonas spp., While the study of36 whose exhibited that 40°C the maximum lipase production for Pseudomonas cepaciea was at 40°C. and the study of37,38 reported that incubation temperature at 50 ̊C was optimum temperature for production of lipase by Bacillus sp. 8°C. The study of39 reported that the showed that the most suitable temperature for the highest lipase activity (225 U/ml) was 10°C, whereas the lowest lipase activity (175 U/ml) was detected at 30°C.
Staphylococcus hemolytic is prosperous bacteria has ability to secrete extracellular lipase. The bacterial isolates from clinical sources were screened as lipase producing bacteria and showing on Rhodamine-B agar plate. Lipase production of organism were measured with varying incubation temperature (20 to 50°C) and incubation time range (16-120)hrs. The present study displayed that Staphylococcus hemolytic (GenBank acc. MG595372.1 ) isolated of the clinical specimens described in high lipase production. The results appeared that the enzymatic activity of crude lipase extract was maximum for incubation period of 72 hours at 37°C about 140 U/ml.
ACKNOWLEDGMENTS
The authors are thankful to Dr. Rafid Mohammed Ali for his efforts and help. We also
like to thank Dr. Ahmed Al–Shameree for his support in genetic part, and thankful to Dr. Ahmed Jalal for Departmental facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HJJA contributed by measurement of lipase activity and the genetic analysis for the isolates. AKA contributed by isolation and identification of the isolates. BMM analyzed
the data and helped for preparation of figures, interpretation and draft.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Panesar PS, Kaur R, Singla G and Sangwan RS. Bio-processing of Agro-industrial wastes for producing of food- grade enzymes: Process and prospects. Appli. Food Biotechnol., 2016; 3(4): 208-227.
- Mahale PK, Desai SV, Hombalimath VS and Achappa S. Isolation, Screening and Characterization of Lipase Producing Strain from Oil Contaminated Soil of Hubballi, Karnataka. Intern. J. Basic and Appl. Bio., 2015; 2(4), pp. 198-201, ISSN: 2394-5820, Online ISSN: 2349-2539.
- Lajis AFB. Realm of thermoalkaline lipases in bioprocess commodities Lajis. Hindawi J. Lipids, 2018, 1–22.
Crossref - Borrelli GM and Trono D. Lipases and Phosphlipases and thier use as biocatalysts for industrial applications. Intern J. Mol Sci., 2015; 16: 20774-20840.
Crossref - Massaoudi A, Belguith H, Gram I and Hamida JB. Classification of E.C.3.1.1.3 bacterial true lipases using phylogenetic analysis. African J. Biotechnol., 2010; 9(48): 8243-8247.
Crossref - Rahman RNZRA, Shariff F, Basri MA and Sallah A. 3D structure elucidation of thermostable L3 lipase from thermophilic Bacillus sp. L2. Intern. J. Mol. Sci., 2012; 13: 9207-9217.
Crossref - Dey A, Maiti TK and Roy P. Improvement of the enzymatic performance of lipase from Pseudomonas sp. ADT3 via entrapment in alginate hydro gel beads. Intern. J. Sci. and Res. Publi., 2015; 5: 1-9.
- Arvidson SO. Extracellular enzymes from Staphylococcus aureus. In Staphylococci and Staphylococcal Infections, 1983; 2: 745-808
- Veerapagui M, Sankara N, Narayanan DM, Ponmurugan K, Jeya K. Screening selection identification production and optimization of bacterial lipase from oil spilled soil. Asian J. Pharma., 2013; 663: 62-67.
- Al-Shuhaib MBS, Al-Kaaby HN, Alwan SL. A highly efficient electrophoretic method for discrimination between two Neoscytalidium species using a specific fungal internal transcribed spacer (ITS) fragment. Folia Microbiologica, 2019; 64(2): 161-170.
Crossref - Zhang Z, Schwartz S, Wagner L, and Miller WA. Greedy algorithm for aligning DNA sequences. J Comput Biol., 2000; 7(1-2): 203-14.
Crossref - Rambaut A. FigTree v1.4.2., 2014. Available at: http://tree.bio.ed.ac.uk/ software/figtree/.
- Akanbi TO, Kamaruzaman AL, Abu Bakar F, Sheikh AN, Radn S, Abul manap MY and Saari N. Highly thermostable extra-cellular lipase producing Bacillus strain isolated from a Malaysian hotspring and identified using 16SrRNA gene sequencing. Inter. Food Res J., 2010; 17: 45-53.
- Rabbani M, Bagherinejad MR, Sadeghi HM, Shariat Z, Rabbari M, Mafakher L and Zaghian S. Isolation and characterization of novel thermophilic lipase-secreting bacteria. Barazil. J. of Microbiol., 2013; 44(4): 1113-1119.
Crossref - Sagar K, Bashir Y, Phukan MM, Konwar BK. Isolation Of Lipolytic Bacteria From Waste Contaminated Soil: A Study With Regard To Process Optimization For Lipase. Intern. J. Sci. and Technol. Res., 2013; 2: 214 – 218.
- Zouaoui B, Bouziane A and Rahoghalem B. Production optimization and purification of lipase from Pseudomonas aeruginosa. Afri. J. of Microbiol. Res., 2012; 6(20): 4417- 4423.
Crossref - Aruna K and Khan K. Optimization studies on production and activity of lipase obtained from Staphylococcus pasteuri SNA 59 isolated from spoit skin lotion. Intern. J. Cur. Microbio. and Appl. Sci., 2014; 3(5): 326-347.
- Bhavani M, Chowdary GV, David M, Archana G. Screening, Isolation and Biochemical characterization of novel lipase producing from soil samples. Intern J. of Biol. Eng., 2012; 2(2).
Crossref - Prasad MP. Evaluation of Growth Parameters on Extracellular Lipase Enzyme Production by Staphylococcus Aureus. International Journal of Science and Research (IJSR), 2015; 4(1).
- Macfaddin JF. Biochemical tests for Identification of Medical Bacteria. 3ed, Lippin cott Williams, Wilkins, Philadelphia, 2000.
- Kukreja V and Bera M. Lipase from Pseudomonas aeruginosa MTCC 2488: Partial purification characterization and calcium dependent thermostability. Ind. J. Biotachnol., 2005; 4: 222-226.
- Boonmahome P and Mongkolthanaruk W. Lipase Producing Bacterium and its Enzyme Characterization. Life Sci. Arch., 2013; 1(4): 335 – 343.
Crossref - Mohammed HJ. Lipase activity of Acinetobacter baumannii 82 Physicochemical Factors Affected the Partial Purified Lipase Activity of Acinetobacter baumannii “local isolates” Iraqi J. Pharm. Sci., 2013; 22(1), P: 82-89.
- Ameri A, Shakibaie M, Amirpour-Rostami S, Ameri A, Adeli-sardou M, Khazali P, Rhahmani H and Forootanfar H. Partial purification and characterization of thermoalkalophilic lipase originated from Bacillus atrophaeus FSHM2 and its application for ester synthesis. Biotechnol. Asian Net. for Scie. infor., 2015; 14(4): 154-164.
Crossref - Peil GHS, Kuss AN, Rave AFG, Villarreal JPV, Hernandes YML and Nascente P. Bioprospecting of lipolytic microorganisms obtained from industrial effluents. Ann. Brazil Acad Sci., 2015; 88: 1769-1779.
Crossref - Alkabbi HJ and AlHamdani MA.Study on the activity and optimization of lipase production bacteria isolated from oil contaminated soil. Indo- Asian J. of Multidisc. Res., 2017; 3(1): 944- 956.
- Salwoom L, Raja Noor Zaliha Raja Abd Rahman RNZR, Salleh A, Shariff FM, Convey P and Ali MSM. New Recombinant Cold-Adapted and Organic Solvent Tolerant Lipase from Psychrophilic Pseudomonas sp. LSK25, Isolated from Signy Island Antarctica. Molecules, 2019; 24(715): 1-14.
Crossref - Kumar A, Parihar S and Btra N. Enrich ment, isolation and optimization of lipase producing Stahpylococcus sp. from oil mill waste (oil cake). Journal of Experament Sci., 2012; 3(8): 26-30.
- Bokhari DM, Gottar UF and Ussain Z. Optimization studies of lipase production from Locally isolated Bacillus spp. Biologia (Pakistan), 2013; 59(2): 295-265.
- Fatima H, Khan N, Rahman AU and Hussain Z. Production and partial characterization of lipase from Pseudomonas putida. Ferment Technol., 2014; 4(1): 2-7.
Crossref - Musa N, Latip W, Abd Rahman RN, Salleh A and Ali MS. Immobilization of an Antarctic Pseudomonas AMS8 lipase for low temperature ethyl hexanoate synthesis. Catalysts, 2018; 8: 234.
Crossref - Subathra Devi SC, Mohanasrinivasan V, Jemimah NS, Yamini B, Chitra M and Nandhini G. Strain improved of Pseudomonas sp. VITSDVM 1for optimization of lipase production by chemical mutagens. Research J. of Pharmaceutical, Biological Chemical Sciences, 2015; 6(3): 782.
- Kulkarni N and Gadre RV. Production and properties of an alkaline, thermophilic lipase from Pseudomonas fluorescence NS2W, J. Ind. Food Microbial., 2002; 28: 344-348.
Crossref - Adan A. Isolation and identification of lipase producing psychrotrophic bacteria from soil: Cloning and partial characterization of its lipase, MSC thesis submitted to the Graduate School of Engineering and Sciences of Izmir Institute of Technology, Izmir university, Turkey, 2009; 1- 60.
- Abdul Faisal P, Hareesh ES, Priji P, Unni KN, Sajith S, Sreedevi S, Josh MS and Benjamin S. Optimization of parameters for the production of lipase from Pseudomonas sp. BUP6 by solid state fermentation. Advances in Enzyme Research., 2014; 2: 125-133.
Crossref - Abdul-Hammid AM, Khedeer MK and Forman MS. Production and purification of lipase from Pseudomonase cepacia and study some effected conditions on production Diyala. Agricultural Sciences J., 2013; 5(2).
- Bhosate HJ, Uzma SZ and Bismile PC. Optimization of lipase production by thermo-alkalophilic Bacillus sp.8C. Research J. of Microbiol., 2015; 10(11): 523-532.
Crossref - Rabbani M, Shafica F, Shayegh Z, Sadeghi HMM, Shariat ZS, Etemadifar Z and Moazen F. Isolation and characterization of a new thermoalkalophilic lipase from soil bacteria. Iranian J. of Pharmaceu Res., 2015; 14(3): 901-906.
- Hassan SWM, Abd El Latif HH, Ali SM. Production of Cold-Active Lipase by Free and Immobilized Marine Bacillus cereus HSS: Application in Wastewater Treatment. Frontiers in Microbiology., 2018; 9: 2377: 1-13.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.