ISSN: 0973-7510
E-ISSN: 2581-690X
After alcoholic fermentation, Oenococcus oeni is the protagonist that carry out malolactic fermentation in order to improve the quality of wine. However, the effectiveness of malolactic fermentation varies according to intraspecific diversity of O. oeni at a genetic level and physicochemical concentrations of intrinsic factors. In this study, we assessed both components by isolating O. oeni from three Chilean vineyards (Maipo, Colchagua y Curicó) and measuring intrinsic components, such as biogenic amines and amino acids, during malolactic fermentation. Both data were condensed and then processed with Multivariate using the statistical method of correspondence analysis. The genetic data was clustered using the RAPD-PCR molecular method, and the physicochemical analysis was carried out using chromatography techniques. Isolation of O. oeni species and genetic diversity results between the vineyards revealed that O. oeni strains cluster according to its geographical origin, with strains similarity higher than 60% of all the samples. The wines of each valley could be characterized by the presence or absence of biogenic amines, and final correspondence analysis showed that there is a differentiation of strains between the studied Valleys, also confirming the accomplishment of the malolactic fermentation in the wines analyzed.
Genotyping, Lactic acid bacteria, Oenococcus, Wine, Fermentation.
Wine is produced by natural alcoholic fermentation of must, which is mostly performed by Saccharomyces yeasts. After alcoholic fermentation, a second fermentation called malolactic fermentation is conducted to improve intensity of flavors and increase microbial stability in the wine2. Malolactic fermentation is carried out by lactic acid bacteria, being Oenococcus oeni the main species in charge of the process3–7. O. oeni is gram-positive bacteria that is able to resist low pH (lower than 3.5), high levels of SO2 (50 mg/L) and ethanol concentration (greater than 10.5 v/v)8. Although O. oeni has good adaptation to wine environment, it has shown phenotypic and genotypic diversity at strain level9 that may have an effect on the result of malolactic fermentation and consequently, it produces inconsistencies in the quality of the wine. This has led to the selection of resistant strains from different environments6, in which intraspecific O. oeni biodiversity studies have been assessed comparing RAPD-PCR results and it has showed a large diversity and correlation between strains and geographical origin10–12. These observations give insights of the potential of identifying O. oeni strains from different geographical regions for the selection of resistant strains and development of fermentation starters that will improve the oenological properties of local wines.
Although malolactic fermentation enhances the wine microbial stability and flavor, the process can be risky because some biogenic amines may increase their levels generating the opposite effect: undesired aroma and flavor13-14. The increased of the concentration of biogenic amines has been attributed to the action of lactic acid bacteria that decarboxylates the amino acids present in the wine, which can also be influenced by intrinsic features such as amino acids concentration, sugars, ethanol concentration, etc.
The aim of this study was to evaluate the effect of genetic diversity on the consumption of amino acids and the production of biogenic amines. For this, we evaluate isolates of O. oeni population genetic clustering obtained of three Central Valleys of Chile: Maipo, Curicó y Colchagua and correlate them with malolactic fermentation, biogenic amines and amino acids concentrations from vintages from the same vineyards using correspondence analysis and Partial Least Square Discriminant Analysis (PLS-DA).
Molecular characterization and O. oeni isolation from Chilean vineyards
Lactic acid bacteria were isolated from 30 Cabernet Sauvignon wine samples from three central Chilean vineyards: Maipo (n=6), Curicó (n=15) and Colchagua (n=9) taken during malolactic fermentation. In addition, seven control strains were used: Pediococcus pentosaceus (CR 949) from Centro de Estudios de Enologia (INTA-Argentina), Lactobacillus plantarum (2793) and Lactobacillus brevis (1366) from Laboratorio de Biotecnología y Microbiología Aplicada (LAMAP, USACH, Chile), Pediococcus acidilactici (B-14950) from USA Agriculture Department, Lactobacillus paracasei (R0212), Lactobacillus hilgardii (H013) and O. oeni (Lalvin 31, Lalvin, France). Serial dilutions method (10-6) was used for isolation, and 100 ìL per sample were grown in basal agar-grape culture media (10 g/L yeast extract, 5 g/L Glucose, 1 mL Tween 80, 170 mL of sterile grape juice, 25 g/L agar) supplemented with cycloheximide 1%. Petri dishes were incubated for 5-7 days at 28 ºC under microaerophilic conditions. Initially, the morphology of cells was observed using Gram tinction15 and the cells with ovoid-shaped and grouped in chains, were preliminarily identified as O. oenioeni bacteria were distinguished at species level using RFLP-PCR method16. First, we extracted total DNA according to kit Wizard DNA purification method (Promega, USA). A fragment of 294 bp that codes for the gene rpoB was amplified, which PCR products were digested with HinfI and AciI digestion enzymes (Fermentas, USA) and separated to be detected at 4% agarose gel electrophoresis.
Samples were labeled as M[A/B/C]-X, which corresponds to molecular information from zone A: Maipo, zone B: Colchagua and zone C: Curicó, and X the isolated strain number. Then, the isolates identified as O. oeni a malolactic region of 1025 bp was amplified with PCR using primers On1 (5’-TAA TGT GGT TCT TGA GGA GAA AAT-3’) and On2 (5’-ATC ATC GTC AAA CAA GAG GCC TT-3’ )17. PCR amplification was carried out during 30 cycles of amplification with denaturation during 45 s at 95°C, annealing for 2 min at 64°C, extension of 2 min at 72°C. PCR products were visualized in 1% agarose gel electrophoresis. Finally, O. oeni strains diversity was evaluated using short primer as previously described RAPD technique10. PCR program was repeated during 30 cycles of amplification with denaturation during 1 min at 94°C, annealing for 2 min at 40°C, extension of 2 min at 72°C, and final 10 min extension at 72°C. Products were visualized in 1.4% agarose gel. All the electrophoresis gels were staining of 5 µg ethidium bromide per L. Electropherograms were processed using the software Quantity One version 4.1.1 (Bio-Rad, USA) to calculate distances employing UPGMA cluster analysis, in order to build a dendrogram with the tool Free Tree versión 0.9.1.5018 and visualized it in Figtree v1.4.2.
Biogenic amines characterization during malolactic fermentation
Wines samples collected during malolactic fermentation were chemically analyzed for: a) Biogenic amines quantification (histamine, tyramine, putrescine, cadaverine and phenylethylamine) from samples collected during malolactic fermentation. For this, was carried out using reversed phase HPLC method, according to previously reported conditions14. Biogenic amines are determined by derivatization using mainly benzoyl chloride and posterior extraction with chloroform. Final residue was resuspended in acetonitrile/water mixture for HPLC analysis. Chromatography separation was carried out at a constant temperature of 22°C using a column of 150 x 4,6 nm of dimensions and 5 µ particle size (LUNA, Phenomenex, USA). b) Amino acids quantification was carried out using reversed phase HPLC method19, the samples were derivatized with phenyl isocyanate in order to form phenylthiocarbamide amino acids, which were separated and quantified through gradient chromatography at a constant temperature of 45 ºC. The mobile phase was composed of a solution (A) of sodium acetate anhydrous (0.14 M, pH 6.4) and acetonitrile (proportion 96:6). And a solution (B) composed of water and acetonitrile (proportion 60:40).
Correspondence analysis between genetic diversity and biogenic amines content
The genetic diversity of O. oeni and the measurements of intrinsic physicochemical parameters of the wine are two independent variables that were collected in the same vineyards. For this reason, both data were compared using the statistical method of correspondence analysis (CA), which aims to compare qualitative data into factorial analysis of both variables. CA is a multidimensional method to conforms homogenous groups from eventual similarities (or differences) of the isolates map indicating relative distance among isolates20. Results of CA were submitted to condensation/reduction of the dimensionality using the method of Karhunen–Loeve21. Following a PLS-DA were performed.
PLS-DA is an asymmetric method to predict one data set separate from another while compute two sets differently. Indeed, PLS-DA is a linear regression method whereby the multivariate variables corresponding to the observations (physicochemical markers) are related to the class membership for each sample. This modeling technique establishes the relationship between two sets of predictor and response variables. It is a correlation analysis that estimates the values of one variable from a set of controllable independent variables14.
The result was validated by full cross-validation routines, minimizing the prediction residual sum of squares function (PRESS) to avoid overfitting the models22. All analyses were performed using the SPSS 19 (IBM SPSS, USA, 2016) and SIMCA P+ 12 (Umetrics AB, Sweden, 2012) software.
Microscopic characterization of isolates
Wine samples collected during malolactic fermentation were analyzed from three Chilean Valleys: Maipo (n=6), Curicó (n=15) and Colchagua (n=9). In total, a 35% of grown bacteria (248 isolates) were randomly selected from petri dishes cultures, which correspond to 63 samples from Maipo Valley, 117 samples from Curicó Valley and 68 samples from Colchagua Valley. Microscopic characterization of isolates revealed that all isolates showed morphologic characteristics associated with O. oeni 8.
Molecular characterization of O. oeni.
After DNA yield and purity was confirmed, results from PCR amplification of O. oeni specie-specific fragments of malolactic enzyme were analyzed. In total, 239 samples showed presence of the 1025 bp fragment (96%) and those with absence (9 samples) were discarded (data not shown). The amplification of the 1025 bp fragment amplifies species-specific primers targeted to the gene encoding the malolactic enzyme of O. oeni17 Therefore, in all the wines analyzed the population of this species was the predominant one. In contrast, Henrriquez-Aedo et al.23 reported that also found Lactobacillus rhamnosus in the samples of Chilean wines analyzed by them. The same authors described that the latter species were endemic to some Chilean winegrowing areas (Limari, Curicó and Itata), which is a species that was not identified in the present study (a case of the Curicó area).
Metadata of those 9 samples revealed that they were taken from wines at the end of alcoholic fermentation in two different valleys, which is in agree with previously reported observations3–8,10
oeni diversity in Cabernet Sauvignon
Out of total 239 O. oeni samples, 32 (13%) were selected from zone A: Maipo, zone B: Colchagua and zone C: Curicó. According to Zapparoli et al.10 method RAPD-PCR clustering analysis showed high diversity and 29 unique samples in four different cluster were identified (Figure 1). Cophenetic correlation of 0.84 indicates good relation between the dendrogram structure and DICE coefficient, but all the sample showed similarity higher than 65%, which has been also reported in Australian wine24. The observed heterogeneity in the samples with respect to the native populations of O. oeni results has been important to study both from the ecological point of view and by possible technological projections. Bon et al. 25 showed a possible association between the presence of oenological sequences and characteristics of interest. So, the study at the molecular level is interesting not only from one point the taxonomic view, but also from ecological and technological aspects.
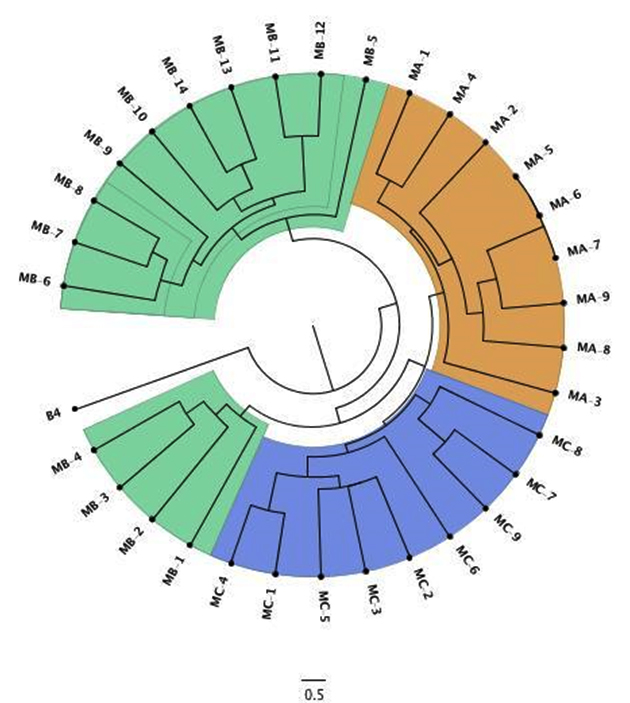
Fig. 1. Dendrogram calculated from UPGMA It corresponds to O. oeni strains molecular information of isolates from three Chilean vineyards. Samples were labeled as M[A/B/C]-X, which corresponds to molecular information from zone A: Maipo, zone B: Colchagua and zone C: Curicó, and then the isolated strain number. B4 is B. bruxellensis outgroup.
Correspondence Analysis (CA)
To identify a relationship between the geographical origin and physicochemical components produced by the strains, we used the CA method linked to PLS-DA. The correspondence analysis revealed that the strains isolated from Colchagua Valley (Figure 2) are grouped, as observed in the RAPD-PCR clustering analysis, supporting the idea of phylogenetic and phenotypic similarity of these strains. It was also possible to appreciate that the Colchagua Valley was characterized by higher levels of lactic acid in its wines, suggesting that malolactic fermentation was performed. Curicó valley was characterized by higher levels of malic acid, which could indicate that the malolactic fermentation was not fully accomplished (Figure 3).
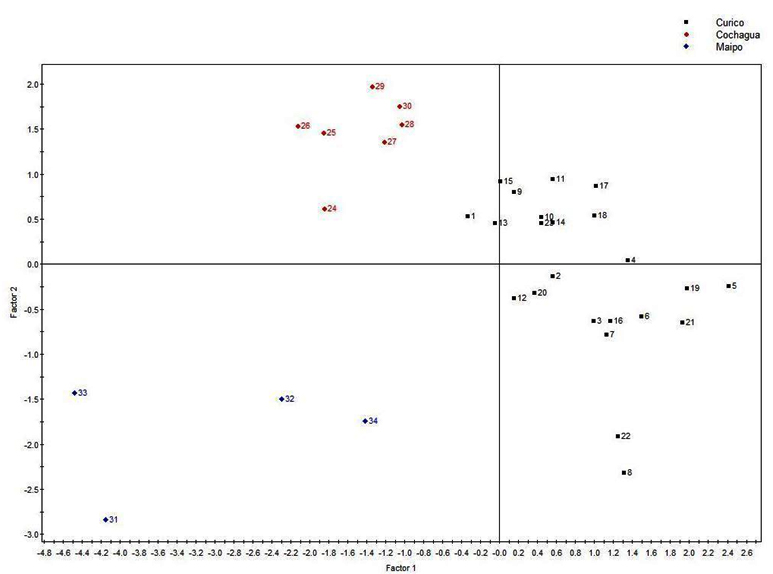
Fig. 2. Partial least squares discriminant analysis (PLS-DA) from HPLC biogenic amines quantification. Samples corresponds to three Chilean Valleys: Curicó (balck: samples 1-23), Colchagua (red: samples 24-30) and Maipo (blue: samples 31-34)
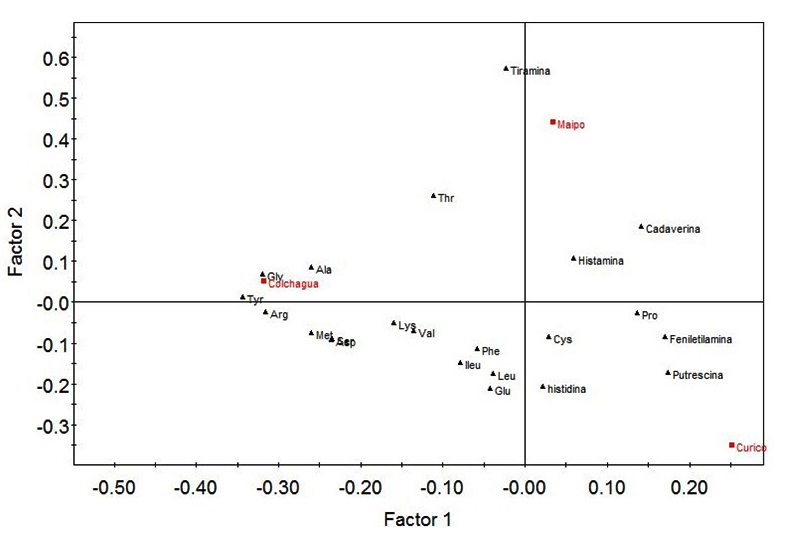
Fig. 3. Partial least squares discriminant analysis (PLS-DA) of molecular information and biogenic amines quantification. The first independent variable corresponds to data from RAPD-PCR results of the genetic variations of O. oeni across three Chilean vineyards: Maipo, Colchagua and Curicó. The second variable corresponds to HPLC quantification of biogenic amines (histamine, tyramine, putrescine, cadaverine and phenylethylamine)
The Maipo Valley, unlike the Curicó and Colchagua valleys, was characterized by the presence of histamine, tyramine and cadaverine. It is important to notice that histamine and tyramine have been suspected to cause toxicological problems26. In our case the levels of this amines were much lower than the physiologically acceptable thresholds27. These biogenic amines are related to microbial activity and their formation occurs mainly in alcoholic and malolactic fermentation28,29. It is thus that this valley (Maipo) showed a low presence of precursor amino acids of the mentioned biogenic amines (Figure 3). On the other hand, the Curicó valley is characterized by the presence of putrescine related with vines suffer stressful conditions30 and phenylethylamine. The presence of the different biogenic amines is related to the amino acid content present in the samples. In this way, the Colchagua valley presents higher levels of amino acids and absence of biogenic amines, which suggests that the bacteria that metabolize malic acid do not metabolized the precursor amino acids associated with biogenic amines. This valley is characterized by the presence of Glycine, Alanine, Tyrosine, Threonine, Arginine, Methionine, Lysine, Valine, among others.
Guerrini et al. 31 determined the biogenic amine-producing capability of O. oeni strains, isolated from different Italian wines, and this capability was quali-quantitatively variable among the strains. Out of the 44 strains investigated, more than 60% of the strains were able to produce histamine, in a range of concentrations between 1.0 to 33 mg/L, and about 16% showed the additional capability to form both putrescine and cadaverine.
Also, Landete et al. 32 found that although O. oeni was the most frequent histamine producer, this species also produced the lowest concentrations and that this production is strain dependent33. Coton et al. 34 reported in a study of 113 wine-isolated O. oeni strains, than 12 strains were positive for the biogenic amine-encoding genes histidine decarboxylase (10 strains) and ornithine decarboxylase (two strains). The ability of one or more strains to predominate in their natural habitat will allow not only to control the production of biogenic amines in wine, but also to deliver properties of oenological interest such as SO2 resistance and sensory characteristics35. Our results showed that biogenic amine-production phenotype is a characteristic strain dependent.
In summary, most of isolated strains from Cabernet Sauvignon were identified as O. oeni. Strain showed similarity higher than 60% in all the samples, but they were clustered according to its geographical origin, indicating adaptation of O. oeni in each Valley and the condition of each vineyard.
ACKNOWLEDGMENTS
This work was supported by grants: Postdoc Dicyt, Código 081671GM PT and USA1555-USACH grant.
- Versari, A., Parpinello, G.P., Cattaneo, M. Leuconostoc oenos and malolactic fermentation in wine: a review. J Ind Microbiol Biotechnol. 1999; 23(6):447–55.
- Siezen, R.J. Wine genomics. Microb Biotechnol. 2008; 1(2):97–103.
- Davis, C.R., Wibowo, D.J., Lee, T.H., Fleet, G.H. Growth and Metabolism of Lactic Acid Bacteria during and after Malolactic Fermentation of Wines at Different pH. Appl Environ Microbiol. 1986; 51(3):539–45.
- Tourdot-Maréchal, R., Cavin, J.F., Drici-Cachon, Z., Diviès, C. Transport of malic acid in Leuconostoc oenos strains defective in malolactic fermentation: a model to evaluate the kinetic parameters. Appl Microbiol Biotechnol. 1993; 39(4-5):499–505.
- Bourdineaud, J.P., Nehmé, B., Tesse, S., Lonvaud-Funel, A. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl Environ Microbiol. 2003; 69(5):2512–20.
- Renouf, V., Vayssieres, L.C., Claisse, O., Lonvaud-Funel, A. Genetic and phenotypic evidence for two groups of Oenococcus oeni strains and their prevalence during winemaking. Appl Microbiol Biotechnol. 2009; 83(1):85–97.
- Solieri, L., Genova, F., De Paola, M., Giudici, P. Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. J Appl Microbiol. 2010; 108(1):285–98.
- Costantini, A., García-Moruno, E., Moreno-Arribas, M. Biochemical Transformations Produced by Malolactic Fermentation. In: Wine Chemistry and Biochemistry (M.V. Moreno-Arribas, M.C. Polo, eds). New York: Springer-Verlag, 2009; pp. 27–57.
- Granchi, L., Guerrini, S., Mangani, S., Vincenzini, M. Biodiversity And Geographical Origin Of Oenococcus Oeni Strains. Acta Hortic. 2007; 754: 147–54.
- Zapparoli, G., Reguant, C., Bordons, A., Torriani, S., Dellaglio, F. Genomic DNA fingerprinting of Oenococcus oeni strains by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA-PCR. Curr Microbiol. 2000; 40(6):351–5.
- Ruiz, P., Izquierdo, P.M., Seseña, S., Palop, M.L. Intraspecific genetic diversity of lactic acid bacteria from malolactic fermentation of Cencibel wines as derived from combined analysis of RAPD-PCR and PFGE patterns. Food Microbiol. 2008; 25(7):942–8.
- Cañas, P.M.I., Pérez, P.R., Prieto, S.S., Herreros, M.L.P. Ecological study of lactic acid microbiota isolated from Tempranillo wines of Castilla-La Mancha. J Biosci Bioeng. 2009; 108(3):220–4.
- Izquierdo-Cañas, P.M., García-Romero, E., Gómez-Alonso, S., Fernández-González, M., Palop-Herreros, M.L.L. Amino acids and biogenic amines during spontaneous malolactic fermentation in Tempranillo red wines. J Food Compost Anal. 2008; 21(8):731–5.
- Yañez, L., Saavedra, J., Martínez, C., Córdova, A., Ganga, M.A. Chemometric Analysis for the Detection of Biogenic Amines in Chilean Cabernet Sauvignon Wines: A Comparative Study between Organic and Nonorganic Production. J Food Sci. 2012; 77(8):T143–50.
- Gunasekaran, P. Laboratory Manual in Microbiology. New Delhi: New Age International, 2007; p152.
- Claisse, O., Renouf, V., Lonvaud-Funel, A. Differentiation of wine lactic acid bacteria species based on RFLP analysis of a partial sequence of rpoB gene. J Microbiol Methods. 2007; 69(2):387–90.
- Zapparoli, G., Torriani, S., Pesente, P., Dellaglio, F. Design and evaluation of malolactic enzyme gene targeted primers for rapid identification and detection of Oenococcus oeni in wine. Lett Appl Microbiol. 1998; 27(5):243–6.
- Pavlícek, A., Hrdá, S., Flegr, J. Free-Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol. 1999; 45(3):97–9.
- Bank, R.A., Jansen, E.J., Beekman, B., te Koppele, J.M. Amino acid analysis by reverse-phase high-performance liquid chromatography: improved derivatization and detection conditions with 9-fluorenylmethyl chloroformate. Anal Biochem. 1996; 240(2):167–76.
- Godoy, L., Garrido, D., Martínez, C., Saavedra, J., Combina, M., Ganga, M.A. Study of the coumarate decarboxylase and vinylphenol reductase activities of Dekkera bruxellensis (anamorph Brettanomyces bruxellensis) isolates. Lett Appl Microbiol. 2009; 48(4):452–7.
- Kroonenberg, P.M. Applied Multiway Data Analysis. New Jersey: John Wiley & Sons, 2008; p 608.
- Cuneo, I., Salgado, E., Castro, M., Cordova, A., Saavedra, J. Effects of climate and anthocyanin variables on the zoning of Pinot Noir wine from the Casablanca Valley. J Wine Res. 2013; 24(4):264–77.
- Henríquez-Aedo, K., Durán, D., Garcia, A., Hengst, M.B., Aranda, M. Identification of biogenic amines-producing lactic acid bacteria isolated from spontaneous malolactic fermentation of chilean red wines. LWT – Food Science and Technology. 2016; 68:183–9.
- Bartowsky, E.J., McCARTHY, J.M., Henschke, P.A. Differentiation of Australian wine isolates of Oenococcus oeni using random amplified polymorphic DNA (RAPD). Aust J Grape Wine Res. 2003; 9(2):122–6.
- Bon, E., Delaherche, A., Bilhère, E., De Daruvar, A., Lonvaud-Funel, A., Le Marrec, C. Oenococcus oeni genome plasticity is associated with fitness. Appl Environ Microbiol. 2009; 75(7):2079–90.
- Bover-Cid, S., Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol. 1999; 53(1):33–41.
- Soufleros, E., Barrios, M.L., Bertrand, A. Correlation Between the Content of Biogenic Amines and Other Wine Compounds. Am J Enol Vitic. 1998; 49(3):266–78.
- Anli, R.E., Bayram, M. Biogenic Amines in Wines. Food Rev Int. 2008; 25(1):86–102.
- Beneduce, L., Romano, A., Capozzi, V., Lucas, P., Barnavon, L., Bach, B., Vuchot, P., Grieco, F., Spano, G. Biogenic amine in wines. Ann Microbiol. 2010; 60(4):573–8.
- Ancín-Azpilicueta, C., González-Marco, A., Jiménez-Moreno, N. Current knowledge about the presence of amines in wine. Crit Rev Food Sci Nutr. 2008; 48(3):257–75.
- Guerrini, S., Mangani, S., Granchi, L., Vincenzini, M. Biogenic amine production by Oenococcus oeni. Curr Microbiol. 2002; 44(5):374–8.
- Landete, J.M., Ferrer, S., Pardo, I. Which lactic acid bacteria are responsible for histamine production in wine?. J Appl Microbiol. 2005; 99(3):580–6.
- Rosi, I., Nannelli, F., Giovani, G. Biogenic amine production by Oenococcus oeni during malolactic fermentation of wines obtained using different strains of Saccharomyces cerevisiae. LWT – Food Science and Technology. 2009; 42(2):525–30.
- Coton, M., Romano, A., Spano, G., Ziegler, K., Vetrana, C., Desmarais, C., Lonvaud-Funel, A., Lucas, P., Cotona, E. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiol. 2010; 27(8):1078–85.
- Malherbe, S., Tredoux, A.G.J., Nieuwoudt, H.H., du Toit, M. Comparative metabolic profiling to investigate the contribution of O. oeni MLF starter cultures to red wine composition. J Ind Microbiol Biotechnol. 2012; 39(3):477–94.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


