ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.2.36 | © The Author(s). 2019
Staphylococcus aureus (S. aureus) is remarkable pathogen and causative agent of hospital acquired (nosocomial) and community acquired infections. S aureus can acquire resistance to most antimicrobial agents (antibiotics). A total 295 S aureus isolates were collected from two local hospitals from various sites of body patients, such as urine, pus, sputum, wound, and blood. Patients’ ages were ranging from 5-84 years from males and females. The number of identified S aureus was 83 detected by traditional and methods and confirmed by molecular method using 16S rRNA gene in polymerase chain reaction (PCR). Antibiotic sensitivity profile was performed to evaluate the resistant and sensitive strains and predominance of methicillin resistant S aureus (MRSA). A total 6 (7.3%) from all S. aureus isolates was MRSA. MRSA isolates were multi-resistant for all antibiotics, which used in the current study. Several genes that participate in antibiotic resistance were investigated. SasX gene was detected in 11 (12.3%) samples which was multi-resistant for many antibiotics. In addition, Furthermore, Exfoliative toxin C and D (etC and etD) genes. The etD gene was detected in 33 (39.75%) samples, but etC gene no detected in all of isolates. In addition, probiotic lactobacilli species (spp.) e.g. L acidophillus, and L reuteri against different isolates of methicillin resistence S aureus from different sites of infections observed variance in their inhibitory activity ranging from 9-17mm. In addition, lactobacilli spp. were collected from different sites and different results showed for S aureus in culture broth and cell free culture supernatant.
Staphylococcus aureus, virulence factors, probiotics, Lactobacillus spp.
Staphylococcu aureus (S. aureus) is one of the pathogens causing a wide range of hospital-acquired (nosocomial) and community-acquired infections. S. aureus play an important role and causative agent of diseases1. The emergence of methicillin-resistant S. aureus (MRSA) was a universal healthcare dilemma due to the limitation of curative options2. At first, it was showed as healthcare facilities specific problem; newly the community-acquired MRSA has raised3. Ness4 predominant that MRSA are resistant to different b-lactam antibiotics but often the MRSA strains are multidrug resistant due to display resistance to other different antibiotics, such as aminoglycosides, macrolides, tetracycline, fluoroquinolones, and chloramphenicol that are usually used in the medication to different diseases caused by these pathogens. Furthermore, in recent two decades S. aureus diseases have become large serious and costly to therapy due to increasing predominance resistivity in S. aureus bacteria due to excessive increase use of antimicrobial agents5. Bimanand et al.6 demonstrated that S. aureus is one of the more common causes of bacteremia and presently carries 20-40% mortality rate monthly. The capability of S. aureus strains to produce adhesion and biofilm makes them more resistant to different antimicrobial agents.
On the other hands, S. aureus is ability forming of biofilm, and increase resistant antibiotics, particularly in the chronic infections those strains forming biofilm and appearance raised resistance against antibiotics treatment are an important medical issue. It is considered which biofilms give a share in more 80% of all infections in human being7.
Furthermore, in S. aureus, there is several virulence factors related with colonization, adherence, and biofilm formation that controlled by polysaccharide intracellular adhesion (PIA)8. SasX is a cell wall-anchored protein that is believed to have contributed to large epidemics of nosocomial infections cause by MRSA in Asia. SasX gene has been displayed to develop S. aureus nasal colonisation in laboratory animals i.e. mouse by encouraging S. aureus adherence to the epithelium of nasal cavity9.
Foster et al.10 stated that the immunization with recombinant SasX gene with specific Abs against SasX decreased colonization for epithelium nasal cavity.
Moreover, exfoliative toxins (ETs) of S. aureus are the major reason of human diseases, particularly staphylococcal scalded skin syndrome (SSSS) in infants and young children. There are three types of ETs; A, B, C, and D. The etA and etB are the main causative agents of SSSS. The etD toxin produces from S. aureus were isolated especially from other cutaneous and soft tissue infections11. Moreover, etC present only in the animals12.
Probiotics defined as live benefit micro-organisms (MO) that confer a health benefits for humans and animals when ingested in sufficient quantities13. Several strains belonging to various MO, such as bacteria and yeast i.e. Lactobacillus, Bifidobacterium, Saccharomyces boulardii, and Saccharomyces cerevisiae, respectively and others, which are the prevalence of the gut microflora14. Probiotics MO together are commensals of the gut and other sites in the body host and differ from MO pathogens in the terms of their actions on the immunity in the gut as they do not stimulate immune system15. Yan and Polk16 stated that the results of evidence based analyses from human being researches and lab animal models have observed the clinical potential of beneficial MO against several diseases. Lactobacilli spp. Produce several substances, such as lactic, and acetic acids, and also to other inhibitory substances e.g. bacteriocins, H2O2, CO2, and others17.
The aims of this study were to investigate the predominant of SasX, etC, and etD genes in S. aureus isolated from samples by polymerase chain reaction (PCR) technique. In addition, estimation effect of some lactobacilli species in vitro by different methods.
Collection of bacterial samples, bacterial maintenance and culturing
A total 295 clinical sample of different infection sites were collected from two local hospitals; Al-Kramah and Al-Zahraa in Wasit city / Iraq. These samples were cultured on the different media, such as blood agar initially and on the selective media i.e. mannitol salt and Staph 110 agars to detect isolates of S. aureus. Bacterial cultures were incubated overnight at 37°C for 18-20 hours (hrs). Other techniques, such as traditional and molecular were used to detect different isolates of S. aureus.
In addition, in the current study, stander probiotic lactobacilli species were used, such as Lactobacillus acidophilus (L acidophilus) ATCC 4356, and Lactobacillus reuteri (L reuteri) ATCC 23272 were prepared and grown on the de Man Rogosa Sharpe (MRS) medium.
Detection of antimicrobial susceptibility test
To conduct of this test, using Bauer-Kirby discs diffusion method to check different isolates susceptibility for different antibiotics. The antibiotics were used in the present study, as the following: Amoxicillin (AMX) 10µg, ampicillin (AMP) 10µg, cefotetan (CTT) 30µg, chlora-mphenicol (CPL) 30µg, ciprofloxacin (CIP) 5µg, clindamycin (CLI) 2µg, Erythromycin (ERY) 15µg, gentamicin (GEN) 10µg, Nitrofurantoin (NIT) 300µg, oxcacillin (OXA) 1µg, tetracycline (TET) 30µg, and vancomycin (VAN) 30µg. Samples were considered as MRSA, according to the oxacillin antibiotic resistance.
StaphylococcuS. aureusDNA extraction protocol and primers with PCR conditions
Genomic DNA in different S. aureus isolates was isolated from S. aureus using Genomic Kit for DNA Extraction (Geneaid Genomic DNA extraction Kit, U.S.A.). DNA was extracted, according to the manufacturer’s instructions. In brief, all samples were centrifuged and pellet was suspended in 0.2 mL GB buffer for 10 minutes. A total 0.2 mL GB buffer was used also for 10 minutes. Then, absolute ethanol (0.2 mL) was added to lysate. 2 ml tube, and used to collect after by GD columns after centrifuge. A W1 buffer added to the GD column and centrifuged. Then, wash buffer was used and elution buffer added and left for 3 minutes to ensure obtaining purified DNA. The different virulence genes responsible for biofilm formation were identified by polymerase chain reaction.
The specific primers, such as 16S rRNA, SasX, etC, and etD were synthesized by Eurofins MWG Operon (MWG, Germany), as outlined in the Table 1.
Finally, concentration of DNA and quality were evaluated using a NanoDrop spectrophotometer and purity. DNA samples with low purity were discarded, and extractions were repeated if required.
Table (1):
All primer used in the current study, and the specific primer of 16S rRNA gene of S aureus
| Primer | Primer’s sequence (5′-3′) | Product size (bp) | Accession No. | |
|---|---|---|---|---|
| SasX | F | TCACCTTTTGCTACACCTGGT | 304bp | KU901576.1 |
| R | AATGACTCAAATGTAAGGGGAGT | |||
| etC | F | TCCCGCACGGTACTTTTGAA | 522 | JX298872.1 |
| R | GAATTATGTCCCAGCCCCGT | |||
| etD | F | AGTACAATGCAGCCCCTTCC | 412 | NC_007795.1:277667 |
| R | CCGGATCCAGAATTTCCCGA | 8-2777550 | ||
| 16S rRNA | F | CTGGAACTGAGACACGGTCC | 777 | KF678862.1 |
| R | CCCAACATCTCACGACACGA | |||
Detection of antagonistic activity using agar spot test method
Overnight cultures of different lactobacilli spp. were spotted onto the surface of MRS agar and incubated for 24hrs. at 37°C to allow colonies to develop. After overnight incubation, the plates were checked for inhibition zones, according to the Klewicka et al.18 method. A clear zone of more than 1 mm around a spot was considered as an indicator of antimicrobial effect19. All tests were replicated three times under the same identical experimental conditions.
Statistical analysis
Statistical analysis was used to analyze data according to the Statistical Package for the Social Sciences (SPSS, Version 17.0). As well as, Chi Square used for all isolates with P value was less than 0.05 (P < 0.05).
Collection of samples
The results of culturing a (295 samples), and taken from different sites of infection in a performance duration extended from February 2018 to February 2019. A total 83 samples of S. aureus were obtained from both sexes; male and female with age range 5-84 years, as listed in the Table 2.
Table (2):
Percentage and numbers of infected patients, according to the age
Age (Year) |
Number |
Percentage (%) |
|---|---|---|
5-14 |
3 |
3.9 |
15-24 |
9 |
14.5 |
25-34 |
12 |
17.1 |
35-44 |
7 |
7.9 |
45-54 |
16 |
14.5 |
55-64 |
21 |
21.1 |
65-74 |
11 |
15.8 |
75-84 |
4 |
5.3 |
Total (Mean) |
83 |
100.0 |
The spread of S. aureus was significant (P < 0.05) variable, according to the gender of infected individual. Females were more than males (54.2% and 45.8%), respectively, as outlined in the Table 3.
Table (3):
Percentage of infected in both; males and females
| Gender | Number | Percentage (%) | |
|---|---|---|---|
| Female | 45 | 54.2 | |
| Male | 38 | 45.8 | |
| Total | 83 | 100.0 | |
P ˂ 0.05
Detection isolates of S. aureus in different samples
S aureus were detected using traditional and molecular techniques. In traditional methods, isolates of S. aureus were ability to produce b-hemolysis on the blood agar medium, mannitol salt fermentation, production of H2O2 from catalase, production of coagulase, and DNase. To confirm diagnosis of S. aureus, detection isolates of S. aureus by molecular technique using 16S rRNA through Maxime Pre Mix for PCR (I NtRON, Korea), according to the manufacturer’s instructions. A total 83 samples were identified as S. aureus bacteria. A total 6 samples were identified as MRSA which constitute 7.3% from all isolates of S. aureus.
According to type of sample, the samples incidence of S. aureus was higher in the pus samples and lower in the burn samples, (39.8% and 6.0%) respectively (P < 0.05), as outlined in the Table 4.
Table (4):
Percentage and numbers of S aureus according to the source of infections
Type of sample |
Numbers |
Percentage (%) |
|---|---|---|
Pus |
33 |
39.8 |
Wound |
15 |
18.0 |
Urine |
11 |
13.3 |
Sputum |
10 |
12.0 |
Blood |
9 |
10.9 |
Burn |
5 |
6.0 |
Total |
83 |
100.0 |
P˂ 0.05
Antibiotic susceptibility test
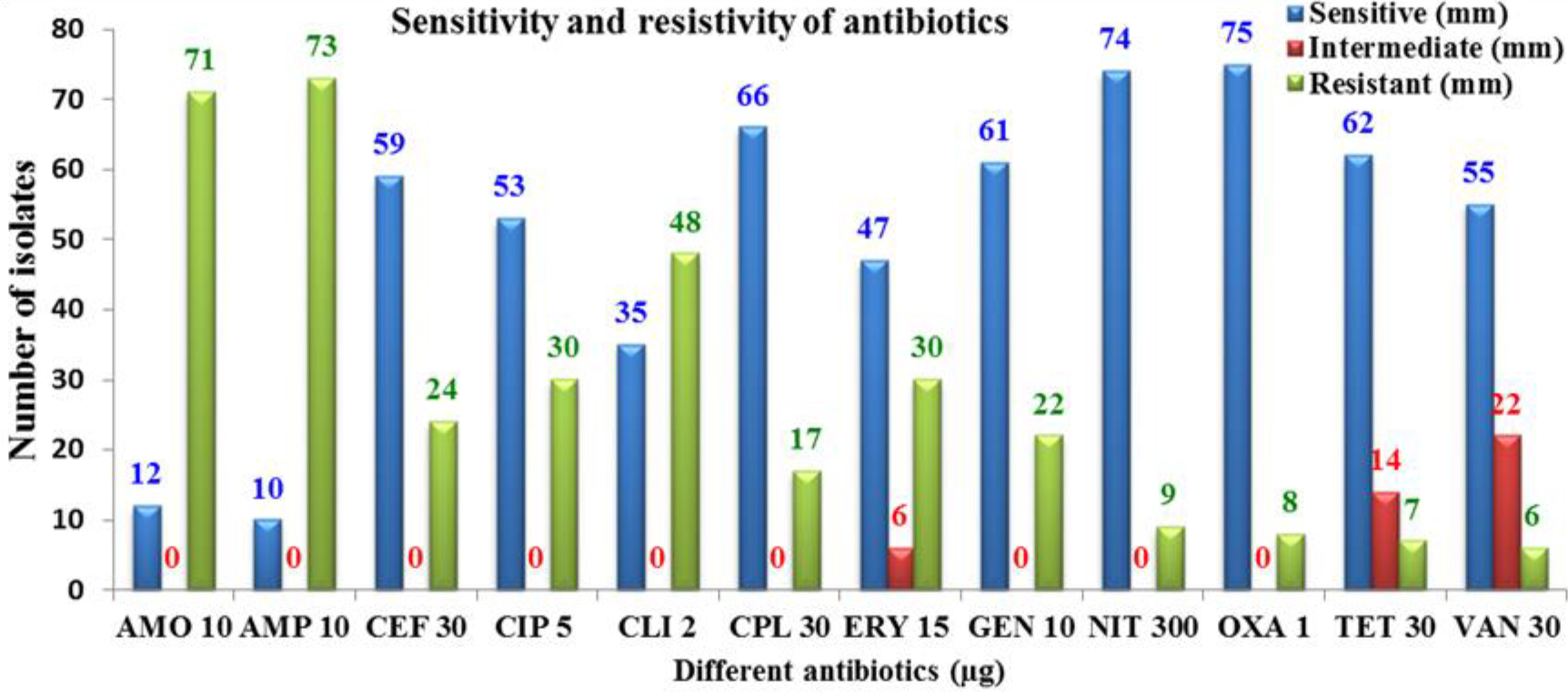
The results for antibiotic susceptibility test were explained in the Fig. 1
Detection of some biofilm and other genes (SasX, etC, and etD) in S. aureus and diagnosis S. aureus isolates by 16S rRNA
Diagnosis of different S. aureus conducted by 16S rRNA (Fig. 2).
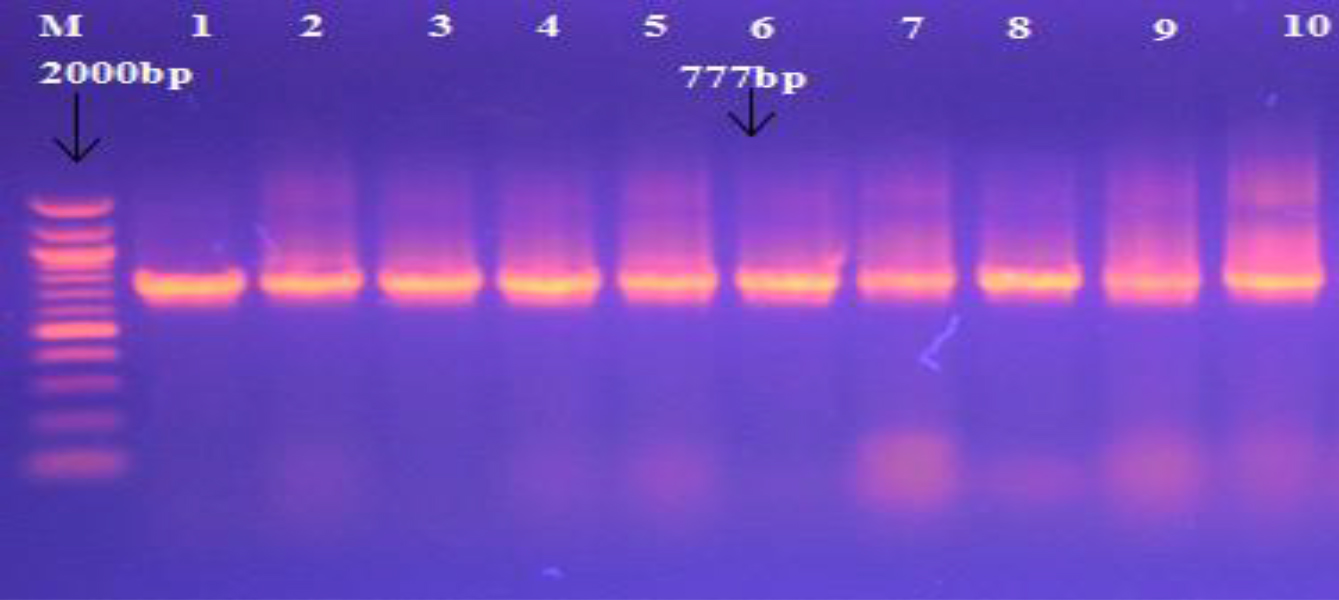
Fig. 2. Agarose gel electrophoresis and Genomic DNA isolated from S. aureus showing 16S rRNA gene for S. aureus using specific primer, with M: Marker (100-2000bp). All lanes were (1-10) positive PCR amplification at 777bp PCR product size.
In addition, PCR was used to detect the presence of different genes, such as exofoliative toxins (etC, and etD) in S. aureus samples. The etD gene was detected in 33 (39.75%) isolates from all S. aureus isolates, as outlined in the Table 5 and the Fig. 3.
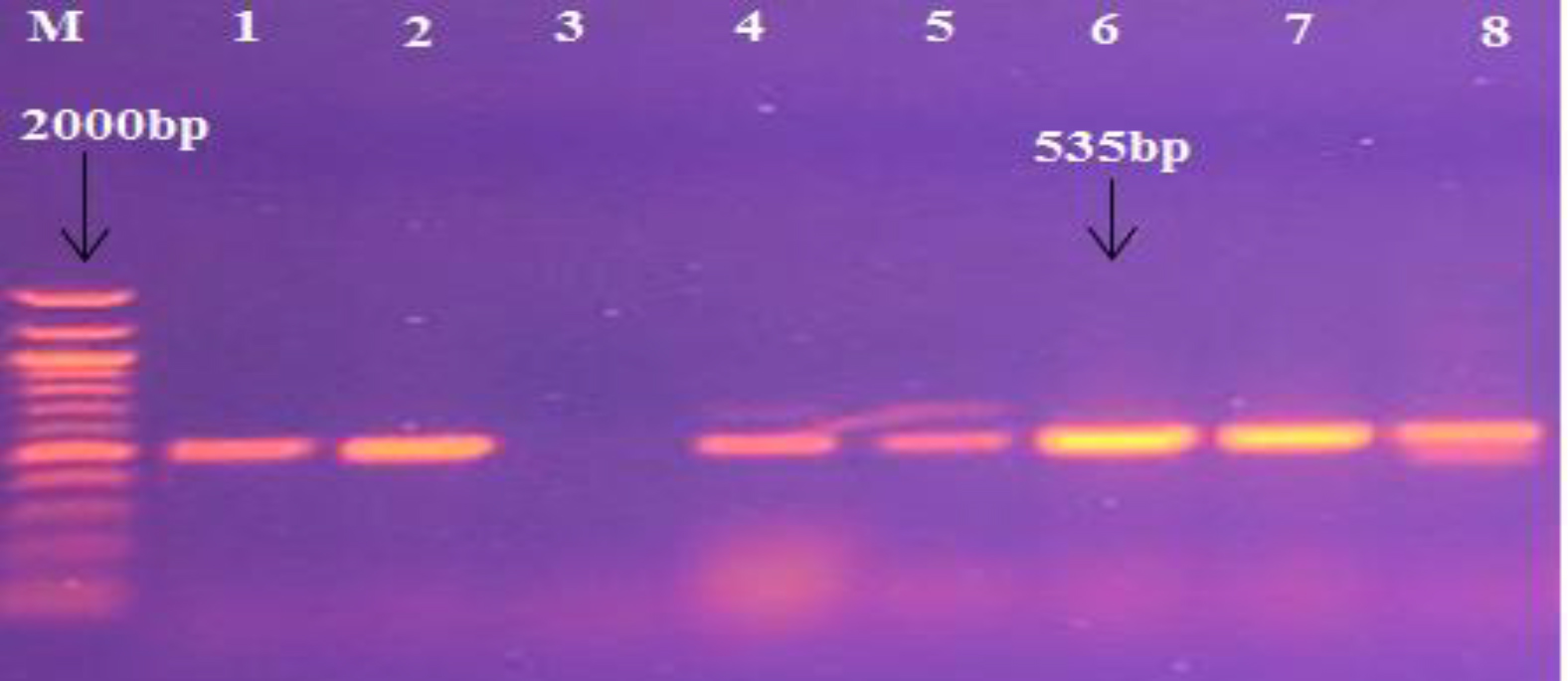
Fig. 3. Agarose gel electrophoresis image that shown the PCR product of etD gene in S. aureus isolates. Where M: Marker (100-2000bp), lane (1-8) some positive PCR amplification at (535bp) PCR product size.
Table (5):
Prevalence of etD gene in different samples of S aureus
| Sample
Gene |
|||||||
|---|---|---|---|---|---|---|---|
| Pus | Wound | Burn | Urine | Sputum | Blood | Total | |
| etD No(%) | 15(45.5) | 10(30.1) | 5(15.2) | 2(6.1) | 1(3.1) | 0(0) | 33(39.75) |
On the other hand, etC gene no detects in all samples of S. aureus samples, as summarized in the Fig. 4.

Fig. 4. Agarose gel electrophoresis image that shown the PCR product of etC gene in S. aureus isolates. Where M: Marker (100-2000bp), lane (1-7) negative PCR amplification at (549bp) PCR product size.
SasX gene in different isolates of S. aureus
Moreover, PCR was used to detect the presence of different genes, such as SasX gene in S. aureus samples. In the current study, the high significant (P < 0.05) rate was in sputum isolates (50%) and lowest was in urine (9.0%) respectively, as outlined in the Table 6 and the Fig. 5.
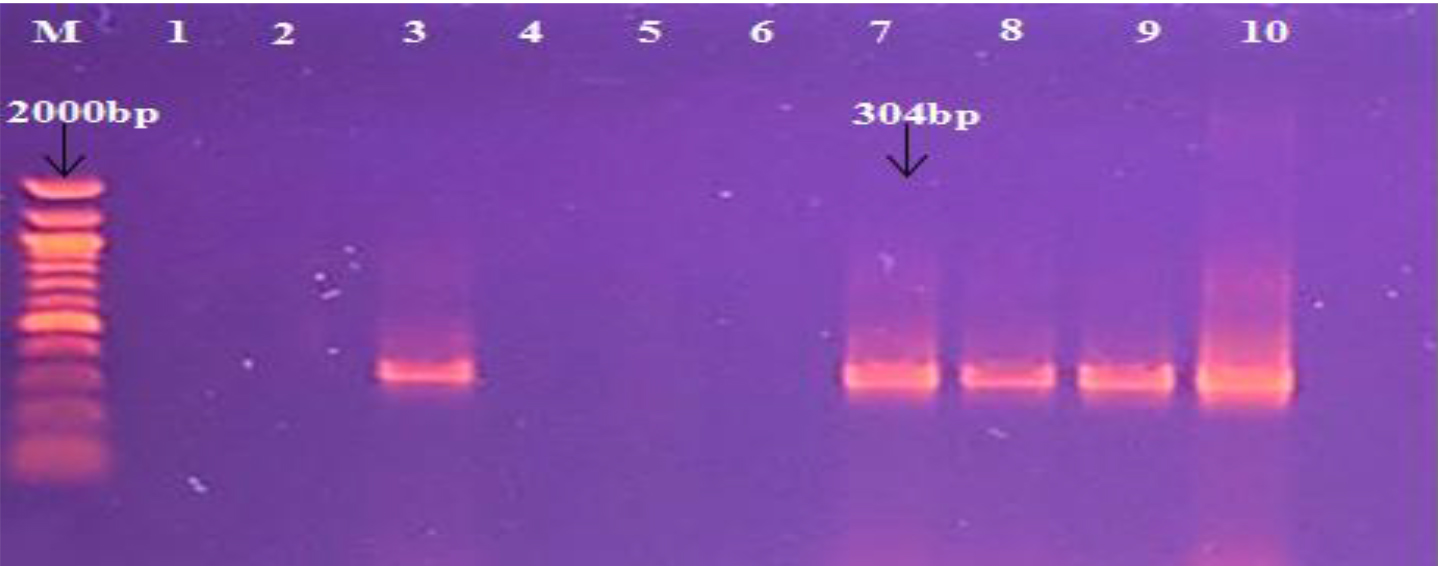
Fig. 5. Agarose gel electrophoresis image that shown the PCR product of SasX gene in S. aureus isolates. Where M: Marker (100-2000bp), lane (1-10) some positive PCR amplification at (304bp) PCR product size.
Table (6):
Prevalence of SasX gene in different samples of S aureus
|
Type |
SasX |
Total No( %) |
||
|---|---|---|---|---|
| Positive No( %) | Negative No( %) | |||
| Pus | 4(12.1) | 29(87.9) | 33(100.0) | |
| Wound | 1(6.7) | 14(93.3) | 15(100.0) | |
| Sputum | 5(50.0) | 5(50.0) | 10(100.0) | |
| Urine | 1(9.1) | 10(90.9) | 11(100.0) | |
| Blood | 0(0.0) | 9(100) | 9(100.0) | |
| Burn | 0(0.0) | 5(100.0) | 5(100.0) | |
| Total No( %) | 11(12.3) | 72(87.7) | 83(100.0) | |
P ˂ 0.05
Assessment inhibitory effect of probiotic lactobacilli species against different S. aureus isolates
The results of lactobacilli species (Standar strains), such as L acidophillus, and L reuteri against different isolates of methicillin resistence S. aureus from different sites of infections observed variance in their inhibitory activity ranging from 9-17mm. Moreover, the results showed that there were variances in the inhibitory effect of the same isolate against antibiotic sensitive and resistant methicillin resistence S. aureus.
Agar spot test method
The results with the current study of bacterial culture broth for Lactobacillus spp. activity test were displayed obvious activity against methicillin resistence S. aureus.
L acidophilus express inhibition zone of 12-17 mm for S. aureus sensitive to antibiotics mentioned above, whereas the results with L reuteri were 9-13 mm, as outlined in the Table 7.
Table (7):
Inhibitory effect of Lactobacillus spp. to methicillin resistence S aureus on the MRS agar.
MRSA number |
L. reuteri (mm) |
L. acidophillus (mm) |
|---|---|---|
1 |
10 |
15 |
2 |
9 |
13 |
3 |
10 |
14 |
4 |
9 |
12 |
5 |
13 |
17 |
6 |
12 |
16 |
S aureusis the major problem which causes a wide spread of diseases, such as bacteremia and nosocomial infections20. Tosas et al.21 demonstrated that S. aureus is has been a worldwide health problem causing high rates of infection and death cases. In the present study, demonstrated there were predictive of high significant variance (P < 0.05) in spread of S. aureus in varies ages of patients which were variable and high percent of infected patients and ranging between 40-59 years.
The majority cases were patients that infected with wound of surgical operations, as summarized in the Table 4. Furthermore, Radhakrishnan et al.22 stated that S. aureus infections distributed between various groups of age similarly, while there were study conducted by Sdougkos et al.23 who reported that the ages between 15-60 years were high susceptible to be infected with S. aureus.
Moreover, the all of results with sensitivity test were significant (P < 0.05). The majority of these S. aureus samples were sensitive to OXA (75mm) and lowest sensitivity was with AMO (12mm), as outlined in the Fig. 1.
Martinez and Baquero24 mentioned that the processes of resistance occur naturally or acquired by mutations or plasmid transportation. On the other hand, the resistance to beta-lactamase was highest with all antibiotics that used in the current study e.g. AMP (73mm) and AMO (71mm) and lowest resistivity was with VAN (6mm), TET (7mm), OXA (8mm), and NIT (9mm), respectively (Table 1). Abdullahi and Iregbu25 stated that the results were resistant as to the occurrence of b-lactamase generating of S. aureus isolates in the health centres environment due to the usage of the b-lactamase treatments for the handling, presenting chance for the selective colonizing for more resistant b-lactamase S. aureus.
With regard to virulence factors, the fact that the existence of SasX gene accompanied with other genes responsible for adherence, colonization, and yield of biofilm and their high resistant to antibiotic, and similar to other researcher like Fahimeh et al.26. The SasX protein has remarkable role in the nasal colonization and epidemic wave of MRSA and effect as a virulence factor on the aggregation of bacteria9. Moreover, the prevalence of SasX isolates were compatible with found Li et al.9 when who reported that SasX gene existence has a role in the enhancing nasal colonization and adhering, abscess formation and lower respiratory tract diseases. The strains of S. aureus which carry SasX gene have been found to spread rapidly in many studies in east China, as reported in Kong et al.27 study. In the present study, 6 (7.3%) were considered isolates as MRSA according to their resistance to OXA, and most of the isolates were isolated from pus and wounds samples. Also, SasX gene has a role in the epidemics of MRSA in Asia, and who stated that SasX gene increase S. aureus colonization, aggregation and evade immune system responses, and lately leads to prolonged survival in blood of human9. In the present study, the percent was higher than results in another study that conducted by Dhiman et al.28 in Shanghai (33.3%).
The etD is a 27 KDa protein and have structural similarity to other exfoliative toxins, was first detected in 2002 by Nishifuji et al.29. The predominant of etD was linked to the cutaneous diseases, and that statement agreed with study which conducted by Mohseni et al.11. The prevalence of etD was linked to the cutaneous diseases, and compatible with found of Bukowski et al.30 study.
Furthermore, Sato et al.12 reported that the molecular mass of another gene of exofoliative C gene (etC) is a 27 kDa, heat labile, and origins etC in the animals, such as chickens and mice. Moreover, etC isolated from horse. In the present study we noticed that there is no presence of etC gene in all samples of human (Fig. 4). These results of current study compatible with found of Nishifuji et al.29 when who reported that etC was not related to human diseases, and identified only in the animal, especially with horse infections.
Probiotic is beneficial intestinal microorganisms have abundant and significant functions, such as they produce several nutrients for body host, inhibit and prevent diseases, which caused by gastrointestinal pathogens, and modulation of immune system response15. For probiotic effect, the results were significant (Table 7) with the current study, and were agreed with found of Dowarah et al.31 who suggested that the inhibitory action of probiotics have main related to statement that lactic acid bacteria (LAB) decrease pH and/or secrete organic acids (lactic and acetic acids). The results of S. aureus culture broth in the current study showed observable inhibitory effect in the resistant and sensitive to isolates of S. aureus. The reason may be due to the production of antimicrobial substances i.e. organic acids which generally the chief end product for LAB. Riviere et al.32 described that the ability to destroy inulin-type fructans of LAB isolates and Prabhujeshwar et al.33 who reported that the ability of organic acids by lactobacilli in various media stated related results. Furthermore, other substances also were noticed to express effect on bacterial growth, such as bacteriocins, and H2O234. The results of the current study were compatible with found of Khanafari and Porgham35, and Vieco-Saiz et al.36.
The percentages of Staphylococcu aureus were increased in pus and wound samples 33 (39.8%) and 15 (18.0%) respectively, and the lowest was in the burn sample 5 (6.0%). Furthermore, MRSA isolates constitute 6 (7.3%) from all S. aureus isolates, and methicillin sensitive S. aureus (MSSA) were 92.7 % from all S. aureus samples. MRSA were multiresistant for numerous antibiotics i.e. ampicillin (73%), amoxicillin (71%) and clindamycin (48%). Furthermore, molecular techneque analysis had seen that the found of adherence, biofilm, and exofoliative genes, especially SasX, etC, and etD, that an increase antibiotic resistance to S. aureus. Moreover, the increase rates of resistance to b-lactam antibiotics in such large rates, and indicates that the misuse of antibiotic represent a serious problem need to be more investigated.
Finally, the current study demonstrated that the probiotic spp had a significant antimicrobial effect against different isolates of S. aureus, especially MRSA, and in several mechanisms and various sources of infections. S. aureus culture broth had a highest effect on the S. aureus than cell-free culture supernatants.
Acknowledgments
Our gratitude and thanks to staff of Al-Karameh and Al-Zahraa hospitals laboratories for their efforts. Moreover, I would like thankful for staff Microbiology Laboratory Departments, College of Medicine, Wasit University for providing requirements and support this work.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Rasigade J.P, Vandenesch F. Staphylococcu aureus: a pathogen with still unresolved issues. Infect. Genet. Evol., 2014; 21: 510-4.
- Jarvis W.R., Jarvis A.A., Chinn R.Y. National prevalence of methicillin-resistant S. aureus in in patients at United States health care facilities, 2010. Am J Infect Control, 2012; 40(3): 194-200.
- Frazee B.W., Lynn J., Charlebois E.D., Lambert L., Lowery D., Perdreau-Remington F. High prevalence of methicillin-resistant S. aureus in emergency department skin and soft tissue infections. Ann Emerg Med., 2005; 45(3): 311-20.
- Ness T. “Multiresistant bacteria in ophthal-mology,” Ophthalmol, 2010; 107(4): 318-322,
- Neyra R.C., Frisancho J.A., Rinsky J.L., et al. “Multidrug-resistant and methicillin-resistant S. aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA),” Environ. Health Perspec., 2014; 122(5): 471-477.
- Bimanand L., Taherikalani M., Jalilian F., Sadeghifard N., Ghafourian S., Mahdavi Z., Mohamadi S., Sayehmiri K., Hematian A., and Pakzad I. Association between biofilm production, adhesion genes and drugs resistance in different SCCmec types of methicillin resistant S. aureus strains isolated from several major hospitals of Iran. Iran J. Basic Med. Sci., 2018; 21(4): 400-403.
- Davies D., “Understanding biofilm resistance to antibacterial agents,” Nature Rev. Drug Discov., 2003; 2(2): 114-122.
- Figueiredo A., Ferreira F., Ossaille C., Cristiana B., Beltrame O., Farrel M., and Marina C. The role of biofilms in persistent infections and factors involved in ica-independent biofilm development and gene regulation in S. aureus. Critic Rev. Microbiol., 2017; 43(5): 1-19.
- Li M., Du X., Villaruz A.E., Diep B.A., Wang D., Song Y., Tian Y., Hu J., Yu F., Lu Y., Otto M. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med., 2012, 18(5): 816-9.
- Foster T.J., Geoghegan J.A., Ganesh V.K., and Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of S. aureus. Nat Rev Microbiol., 2014; 12(1): 49-62.
- Mohseni M., Rafiei F., and Ghaemi E.A. High frequency of exfoliative toxin genes among S. aureus isolated from clinical specimens in the north of Iran: Alarm for the health of individuals under risk. Iran J. Microbiol., 2018; 10(3): 158-165.
- Sato, H., Matsumori, Y., Tanabe, T., Saito, H., Shimizu, A., Kawano, J. A new type of staphylococcal exfoliative toxin from a S. aureus strain isolated from a horse with phlegmon. Infect. Immun., 1994, 62: 3780-3785.
- Food and agriculture organization of the United Nations (FAO). Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria, 2001.
- Bermudez-Brito M. Plaza-Dםaz J. Muסoz-Quezada S. Gףmez-Llorente C. Gil A. Probiotic Mechanisms of Action. Ann. Nutr. Metab., 2012; 61: 160-174.
- Markowiak P., and Slizewska K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients., 2017; 9(9): 1021.
- Yan F., Polk D.B. Probiotics and immune health. Curr. Opin. Gastroenterol., 2011; 27: 496-0501.
- Ogunbanwo, S. Functional properties of lactic acid bacteria isolated from ogi and fufu, two Nigerian fermented foods. Adv. Food Sci., 2005; 27: 14-21.
- Klewicka E., Libudzisz Z., Czajka D., Kuc K. Antagonistic activity of lactic acid bacteria Lactobacillus acidophilus. [Antagonistyczna aktywno ז bakterii fermentacji mlekowej Lactobacillus acidophilus.] Zywnosc 1999; 4(21): 168-175.
- Tahara, T., Oshimura, M., Umezawa, C., and Kanatani, K. Isolation, partial characterization, and mode of action of Acidocin J1132, a two-component bacteriocin produced by Lactobacillus acidophilus JCM 1132. Appl. Environ. Microb., 1996; 6(2): 892-897.
- Christoph and Naber. S. aureus Bacteremia: Epidemiology, Pathophysiology, and Management Strategies. Clin. Infect. Dis., 2009, 48, 4(15): S231-S237.
- Tosas Auguet O., Stabler R.A., Betley J., et al. Frequent Undetected Ward-Based Methicillin-Resistant StaphylococcuS. aureus Transmission Linked to Patient Sharing Between Hospitals. Clin. Infect. Dis. 2018; 66: 840.
- Radhakrishnan V., Ravichandran S., Murugaiyan S., Vadivelu., and Radhakrishnan S. Prevalence of carriage of community-acquired Methicillin-resistant StaphylococcuS. aureus among children attending the pediatric OPD at a tertiary care hospital. Int. J. Contemp. Pediatr., 2018; 5(5): 1956-1961.
- Sdougkosa G., Chini V., Papanastasiouc D.A. Christodouloua G. Stamatakisa E., Vrisa A., Christodoulidid I., Protopapadakisa G., and Spiliopoulou I. Community-associated S. aureus infections and nasal carriage among children: molecular microbial data and clinical characteristics. Clin. Microbiol. Infect., 2008; 14(11): 995-1001.
- Martinez J.L., and Baquero F. Mutation Frequencies and Antibiotic Resistance. Antimicrob agents chemo,. 2000; 44(7): 1771-1777.
- Abdullah N., and Iregbu K.C. Methicillin Resistant StaphylococcuS. aureus in a Central Nigeria Tertiary Hospital. Ann. Tropic. Pathol., 2018; 9(1): 6-10.
- Fahimeh, N., morteza, A., Namvar, E. Detection of genes involved in biofilm formation in S. aureus isolates. GMS. Hygiene Infec. Control, 2016; 11: ISSN 2196-5226.
- Kong H., Fang L., Jiang R., and Tong J. Distribution of SasX, pvl, and qacA/B genes in epidemic methicillin-resistantS. aureus strains isolated from East China. Infect Drug Resist., 2018; 11: 55-59.
- Dhiman M., Bhatt N., Chau-Han V., Farooq A., Khan A. Emergence of methicillin resistant S. aureus (MRSA) isolates from north India harboring a novel SasX gene: further analyzing its role in biofilm formation. Ann Med Biomed Sci., 2017; 3(1): 46-50.
- Nishifuji, K., Sugai, M., Amagai, M. Staphylococcal exfoliative toxins: molecular scissors of bacteria that attack the cutaneous defense barrier in mammals. J. Dermatol Sci., 2008; 49: 210-231.
- Bukowski M., Wladyka B., and Dubin G. Exfoliative Toxins of StaphylococcuS. aureus. Toxins (Basel). 2010; 2(5): 1148-1165.
- Dowarah R., Verma A.K., Agarwa N., Singh P, Singh B.R. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE, 2018; 13(3): e0192978.
- Riviere A., Selak M., Geirnaert A., Abbeele P.V., Vuyst L.D. Complementary Mechanisms for Degradation of Inulin-Type Fructans and Arabinoxylan Oligosaccharides among Bifido-bacterial Strains Suggest Bacterial Cooperation. Appl. Environ. Microbiol., 2018; 84(9): e02893-17.
- Prabhurajeshwar C., and Chandrakanth R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. J. Biomedical., 2017; 40(5): 270-283.
- Gaspar C., Donders G. G., Palmeira-de-Oliveira R., Queiroz J. A., Tomaz C., Martinez-de-Oliveira J., and Palmeira-de-Oliveiracorresponding A. Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Express. 2018; 8: 153.
- Khanafari A., and Porgham S.H. Investigation of Probiotic Chocolate Effect on Streptococcus mutans Growth Inhibition. Jundishapur J Microbiol., 2012; 5(4): 590-597.
- Vieco-Saiz N., Belguesmia Y., Raspoet R., Auclair E., Gance F., Kempf I., Drider D. enefits and Inputs From Lactic Acid Bacteria and Their Bacteriocins as Alternatives to Antibiotic Growth Promoters During Food-Animal Production. Front. Microbiol. 2019 | https://doi.org/10.3389/fmicb.2019. 00057.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.