ISSN: 0973-7510
E-ISSN: 2581-690X
Carbapenem resistance is increasing and emerging as a public health threat. The Indian subcontinent serves as a reservoir for all 3 types of carbapenemases: KPC, OXA-181 and NDM. The present study was done to determine the antibiotic resistance pattern of Gram negative bacilli (GNB) belonging to family Enterobacteriaceae and molecular detection of blaKPC gene among the Carbapenem resistant Enterobacteriaceae (CRE). Antibiotic sensitivity pattern of 301 gram negative isolates, members of Enterobacteriaceae (E. coli, K. pneumoniae, K. oxytoca, Proteus species, Citrobacter species, Enterobacter species and Serratia species) was determined and those resistant to carbapenem (ertapenem) were further processed in the study. Modified Hodge Test (MHT) was performed to phenotypically confirm the presence of carbapenemases (blaKPC) and Polymerase Chain Reaction (PCR) was done to identify blaKPC gene. Of the total 301 isolates, 45 were resistant to ertapenem. Out of these 45 isolates 32 were MHT positive and three of these 32 MHT positive strains harboured blaKPC gene.
blaKPC, Enterobacteriaceae, Gram negative bacilli, Carbapenem resistance, Modified hodge test.
The members of family Enterobacteriaceae are Gram negative bacilli (GNB) which are a part of human normal flora. They are also a source of community and hospital acquired infections1.
Carbapenems belong to a group of beta lactam antibiotics which have a broad spectrum activity. They are being used for treatment of severe life threatening infections caused by Multidrug resistant (MDR) GNB. These agents have become ineffective due to misuse and development of resistance against them2.
Resistance has been developing against the antibiotics soon after they have been put to use. Alexander Fleming the discoverer of penicillin, while accepting his Noble Prize in 1945 warned about the possibility of drug resistance. Resistance to carbapenems was first demonstrated in the North Carolinan strain of Klebsiella pneumoniae that produced an enzyme Klebsiella pneumoniae carbapenemase (KPC). The gene coding for resistance was found on plasmid. It was then when carbapenem resistance arrived3.
KPCs are beta lactam enzymes produced by GNB (K. pneumoniae, E. coli, Enterobacter species) belonging to Enterobacteriaceae family harboring blaKPC gene. These enzymes hydrolyze and inactivate a wide range of beta lactam antibiotics such as penicilllins, cephalosporins and carbapenems. There are different variants of blaKPC gene (KPC-2 to KPC-15) which are seen in non fermenting bacteria also4.
In order to contain the spread of bacteria harboring blaKPC gene effective control measures and judicious antibiotic usage in hospitalized patients must be implemented.
The study focused on KPC producers because they are an important resistance mechanism for wide range of GNB which is not only limited to K. pneumoniae. KPC producing isolates can be misidentified by routine tests and should be suspected by ertapenem resistance. The study was done to identify blaKPC and not other carbapenemases as no other study has been done previously to detect blaKPC gene in a hospital situated in budhera village in Haryana.
The study was conducted in the microbiology laboratory of SGT Medical College and Hospital for a period of 8 months (March-October 2018) on the patient samples received from various clinical departments.
The institutional ethical committee approval was taken prior to commencement of this study.
The GNB isolated from various clinical samples like blood, sputum, pus, urine, vaginal swabs, urinary catheter tip were included for the study. All the samples were inoculated on MacConkey and blood agar except urine samples for which cysteine lactose electrolyte deficient (CLED) agar was used. The organisms were identified by Gram staining and biochemical tests (Indole test, Methyl red, Voges Proskauer, Citrate utilization test, Urea hydrolysis test and Triple Sugar Iron agar test) were done. Antibiotic susceptibility testing was done by Kirby bauer disc diffusion method as per CLSI guidelines 20185. Antimicrobial discs used were ampicillin (10µg), ampicillinsul bactum(10/10µg), ceftazidime (30µg), cefuroxime (30µg), cefotaxime(10µg), cefepime(5µg), levofloxacin (5µg), ciprofloxacin (5µg), tobramicin (10µg), cotrimoxazole (1.25/23.75µg), gentamicin (10µg), ertapenem(10µg), meropenem(10µg), imipenem (10µg), aztreonam (30µg). Resistance to ertapenem served as a screening test for blaKPC gene detection5.Out of 301 isolates, 45 isolates resistant to ertapenem were further processed for Modified Hodge Test and detection of blaKPC gene.
Phenotypic Test for detection of Carbapenemases
Modified Hodge Test (MHT)
Mueller hinton agar (MHA) plate was inoculated as a lawn culture with 0.5 Mc Farland E. coli ATCC 25922. The plate was dried for 3 to 10 minutes and ertapenem disc was applied on the centre of the plate. Using a 10-μL loop, 3 to 5 colonies of test and Quality Control organism which were grown overnight on a blood agar plate were inoculated in straight lines perpendicular to each other. After overnight incubation, the plate was looked for the presence of a “clover-leaf’’ indentation in the zone of inhibition. The isolates showing clover-leaf like indentation around the zone of inhibition of E. coli ATCC 25922 were considered as positive and absence of indentation was considered as negative. Quality control strains used were:
- K. pneumoniae ATCC BAA- 1705 (MHT positive)
- K. pneumoniae ATCC BAA-1706 (MHT negative).
blaKPC gene detection
DNA Extraction, PCR and Electrophoresis were performed according to Shanmugam et al6 with minor modifications. The following primers were used to amplify the blaKPC gene: Forward Primer: 5’-GCT CAG GCG CAA CTG TAA G-3’ and Reverse Primer: 5’-AGC ACA GCG GCA GCA AGA AAG-3’.
PCR Reaction
To make 25µl reaction mix. 12.5µl of HiChrom PCR Master Mix was added. To it 0.75µl of Forward Primer, 0.75µl of Reverse Primer, 2.5µl of Template DNA and 8.5µl of Molecular Biology Grade water was added.
The PCR cycles followed were:
- Initial denaturation was done at 94oC for 3 minutes.
- It was followed by 30 cycles of denaturation at 94oC for 1 minute.
- Annealing was done at 60oC for 1 minute.
- Extension at 72oC for 1 minute which was followed by final extension for 5 minutes at 72oC. Gel electrophoresis was performed on 2% agarose gel at 100volts for 15 minutes. The gel was observed under UV Transilluminator.
During the study period, 301 isolates of Enterobacteriaceae (E. coli, K. pneumoniae, K. oxytoca, Proteus spp., Serratia spp., Citrobacter spp., Enterobacter spp.) were tested by Kirby bauer disc diffusion method, of which 45(14.9%) were found to be Carbapenem Resistant Enterobacteriaceae (CRE).Citrobacter spp., Enterobacter spp. and Serratia spp. were susceptible to Ertapenem, so they were not processed further for Phenotypic and Genotypic tests. All (100%) of CRE were resistant to ertapenem according to CLSI 2018 guidelines. Of the 45 ertapenem resistant isolates, 23(51.1%) were from urine, 16(35.5%) from pus, 3(6.6%) from sputum and 3(6.6%) from blood. Of the forty five isolates twenty nine were E. coli, twelve were K. pneumoniae, three K. oxytoca and one Proteus mirabilis.
Table (1):
Co-resistance pattern of ertapenem resistant isolates.
Antibiotics |
Klebsiella(n5=15) |
E.coli(n1=29) |
Proteus(n1=1) |
|---|---|---|---|
Ampicillin |
15 (100%) |
29 (100%) |
1 (100%) |
Ampicillin sulbactum |
15(100%) |
29(100%) |
1 (100%) |
Ceftazidime |
15(100%) |
29 (100%) |
1 (100%) |
Cefuroxime |
15 (100%) |
29 (100%) |
1 (100%) |
Cefotaxime |
15(100%) |
29 (100%) |
1 (100%) |
Levofloxacin |
15(100%) |
28 (96.5%) |
1 (100%) |
Cefepime |
15(100%) |
29 (100%) |
1 (100%) |
Ciprofloxacin |
15(100%) |
29(100%) |
1 (100%) |
Tobramicin |
11(73.3%) |
21 (72.4%) |
0% |
Gentamicin |
11(80%) |
23 (79.3%) |
1 (100%) |
Cotrimoxazole |
14 (93.3%) |
27 (93.1%) |
0% |
Aztreonam |
11(73.3%) |
22 (75.8%) |
0% |
Norfloxacin |
4 (100%) |
28 (96.5%) |
1 (100%) |
Nitrofurantoin |
3 (75%) |
20 (70%) |
0% |
Imipenem |
14 (93.3%) |
21 (72.4%) |
1 (100%) |
Meropenem |
10(66.6%) |
22 (75.8%) |
0% |
Ertapenem |
15(100%) |
29 (100%) |
1 (100%) |
n5 – no. of isolates
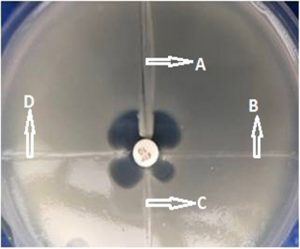
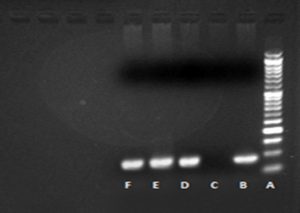
Of the 45 CRE, 32 (71.1%) were resistant to meropenem, 36 (80%) were resistant to imipenem. The resistance pattern of these isolates is given in table 1. All carbapenem (ertapenem) resistant isolates were subjected to Modified Hodge Test. MHT was positive for 32(71.1%) of 45 ertapenem resistant isolates confirming the presence of significant carbapenems hydrolyzing activity (Fig. 1). PCR for detection of blaKPC gene was carried out for MHT positive isolates. Three (9.3%) of the 32 isolates were found to harbor blaKPC gene. Band formation was seen at 90 basepair as shown in Fig. 2. All the KPC positive isolates were K. pneumoniae and they showed resistance to both meropenem and imipenem. Two of the blaKPC isolates were isolated from blood and one from pus sample from patients admitted in Surgery and ICU ward. Twenty nine (90.6%) of 32 MHT positive isolates were blaKPC negative.
Fig. 1. Modified Hodge Test (A is Negative control, B is Positive control, C and D are two test isolates showing positive MHT)
There are reports of increased prevalence of carbapenems resistant GNB over few years. The rapid dissemination of Carbapenemases like KPC is a major challenge for physicians and clinical laboratories. However, identifying the different bacterial mechanisms of resistance is crucial for infection control and epidemiological studies7. KPC producing isolates have been reported from USA, Italy, China, Greece, Argentina, Israel, Poland, Taiwan and Columbia. There are reports of sporadic occurrence of KPC from South East Asia including India, Australia and South Korea8.
Developed countries like United States reported 62.3% CRE in the year 2012-2013. Greece reported <1 % CRE in 2001 which in 2008 rose to 30 % in hospital wards and 60% in ICUs9.
However, various studies conducted in India before 2006 failed to show any evidence of resistance for E. coli and K. pneumoniae to carbapenems10, Indian subcontinent serves as a reservoir for all 3 types of carbapenemases: KPC, OXA-181 and NDM8.
Two different studies from Western Rajasthan reported 31.7% and 37% CRE respectively11, 12. Whereas, the present study showed 14.9% CRE, this is lesser than the other two studies. However, it is in accordance to a study conducted in a Tertiary Care Hospital, North India15 which showed 14.7% CRE.
In the present study, CRE isolates exhibited increased prevalence of multidrug resistance to various antibiotics ranging from 66.6% to 100% which is comparable to a study from Northeast India with resistance rate ranging from 85.7% to 100%14.
MHT was positive in 32 (71.1%) isolates, whereas, only 3(9.3%) showed the presence of blaKPC gene by conventional PCR. However, this non specificity of MHT was discussed by Tsakris et al15.Their report states that the false positive results of MHT could be due to presence of CTX-M ESBL positive or AmpC–hyperproducing Enterobacteriaceae. This also suggests the involvement of other resistance mechanisms, such as production of carbapenemases (OXA, NDM, MBL) alone or in combination with porins loss, ESBL (TEM, SHV, CTX-M)7 .
KPC producing strains in the current study were 100% resistant to ertapenem, imipenem and meropenem. The first report of KPC producer was from South India (Pondicherry) in 201016. It reported six (5.8%) out of 103 isolates as blaKPC positive, that were 100% resistant to carbapenem (imipenem, ertapenem and meropenem).
However, this is the first study conducted in SGT medical college and hospital to detect the prevalence of blaKPC gene.
The present study reported 9.3% KPC isolates which is more than the findings of a study conducted at Safdarjung Hospital, New Delhi 17 that reported 2.1% KPC producers. However, absence of blaKPC was reported in a study conducted in North India by Mohan et al 18 that is contrary to the present study. Although, there are a few studies from India that shows the prevalence of CRE in clinically relevant isolates but not many laboratories follow the standard protocol for CRE identification.
The emergence of carbapenem resistance globally and in India is a cause of concern because of the limited treatment options for carbapenem resistant organisms. More studies should be undertaken in different regions of the country to avail the information on prevalence of KPC. Early diagnosis of KPC can improve the patient outcome. Whereas, the diagnostic technique to be introduced in the laboratory workflow is a choice which should be done carefully, as per the resources available and personnel in each hospital.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AS conducted the isolation and identification of bacterial strains, performed the antibiotic sensitivity tests, phenotypic and genotypic tests, tabulation of data and drafting of the manuscript. PA and MM guided in the phenotypic and genotypic test and correcting the manuscript. AC guided in the final editing of the manuscript. All the authors read and approved the final manuscript.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets created or investigated during this study are involved in the manuscript.
- Nordmann P, Naas T, Poirel L. Global Spread of Carbapenemase- producing Enterobacteriaceae. Emerging Infectious Diseases, 2011; 17(10):1791-98.
Crossref - Ramadan RA, Gebriel MG, Kadry HM et al. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infection and Drug Resistance.2018;11: 1261–69.
Crossref - Maryn Mckenna. Nature 2013; 499:394-96.
Crossref - Cristina P, Ribeiro S , Monteiro AS et al. Phenotypic and molecular detection of the blaKPC gene in clinical isolates from inpatients at hospitals in Sao Luis. BMC Infectious Diseases.2016; 16:737.
Crossref - Clinical Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-eighth Informational Supplement. M100-S28, CLSI. Wayne, Pennsylvania, USA. 2018.
- Shanmugam P, Meenakshisundaram J, Jayaraman P. blaKPC gene Detection in Clinical Isolates of Carbapenem Resistant Enterobacteriaceae in a Tertiay Care Hospital. Journal of Clinical and Diagnostic Research.2013; 12 :2736-38.
Crossref - Biberg CA, Rodrigues ACS, Carmo SF et al. KPC-2 producing Klebsiella pneumoniae in a hospital in Mid West region of Brazil. Journal of Microbiology 2015;46(2):501-504.
Crossref - Remya P, Mariappan Shanthi, Sekar U. Prevalence of blaKPC and its occurrence with other beta lactamases in Klebsiella pneumoniae. J Lab Physicians 2018; 10(4): 387–91.
Crossref - Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis. 2017 ;215: 28–36.
Crossref - Grover SS, Doda A, Gupta N et al. New Delhi metallo-β- lactamase-type carbapenemases producing Escherichia coli isolates from hospitalized patients: A pilot study. Indian J Med Res 2017; 146: 105-10.
Crossref - Pawar SK, Mohite ST ,Shinde RV et al. Carbapenem-resistant Enterobacteriaceae: Prevalence and bacteriological profile in a tertiary teaching hospital from rural western India. Indian Journal of Microbiology Research 2018; 5(3):342-47.
Crossref - Rajni E, Rajpurohit V, Rathore P et al. Epidemiology of carbapenems-resistant Enterobacteriaceae colonization in ICU: a pilot study from a tertiary care hospital in Western Rajasthan, India. Int J Res Med Sci. 2018; 6(10):3340-3345.
Crossref - Dahiya S, Singla P, Chaudhary U et al. Prevalence of Klebsiella pneumoniae Carbapenemase (KPC), Metallo Beta Lactamases and AmpC beta Lactamases in Clinical Isolates of Klebsiella Species. Int.J.Curr.Microbiol.App.Sci. 2015; 4(9): 170-76.
- Bora A , Ahmed GU, Hazarika NK et al. Incidence of bla NDM-1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol. 2013; 31(3):250-56.
Crossref - Tsakris A, Themeli-Digalaki K, Poulou A et al. Comparative Evaluation of Combined-Disk Tests Using Different Boronic Acid Compounds for Detection of Klebsiella pneumoniae Carbapenemase-Producing Enterobacteriaceae Clinical Isolates. Journal of Clinical Microbiology 2011; 49(8): 2804-09.
Crossref - Parveen RM, Harish BN, Parija SC. Emerging Carbapenem Resistance among Nosocomial Isolates of Klebsiella Pneumoniae in South India. International Journal of Pharma and Bio Sciences 2010; 1(2).
- Mohanty S, Mittal G, Gaind R. Identification of carbapenemase mediated resistance among Enterobacteriaceae in bloodstream isolates: A molecular study from India. Indian J Med Microbiol. 2017; 35:421-25.
Crossref - Mohan B, Hallur V, Gagandeep Singh G et al. Occurrence of blaNDM-1 & absence of blaKPC genes encoding carbapenems resistance in uropathogens from a tertiary care centre from north India. Indian J Med Res. 2015; 142(3): 336–43.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.