ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-resistant Staphylococcus aureus (MRSA) strains are widespread globally. Besides their virulence factors, the co-occurrence of antimicrobial and metal resistance has been reported. This study was designed to evaluate the antibiotic resistance and resistance phenotypes, investigate the occurrence of virulence factors, and detect heavy metal tolerance among MRSA strains. Antibiogram profiling was done as recommended by CLSI instructions. Resistance phenotypes were detected by D test, followed by characterization of enzymatic activities and biofilm formation assay. Antibacterial activity of different heavy metals was tested, and predictable synergistic assay was performed. Among MRSA strains collected, high resistance to ampicillin and amoxicillin/clavulanate (100%) and high susceptibility to clindamycin (70%) were obtained. Resistance phenotypes were detected as S, constitutive MLSB, inducible MLSB, and MS by percentages of 10%, 30%, 30% and 30% respectively. Virulence factors like lipolytic (50%) and hemolytic (70%) activity, and biofilm formation ability (100%) were detected. High resistance towards potassium and magnesium was observed. MTC of 500 ppm was detected for all isolates in case of cobalt and iron. In case of zinc and copper, MTC was detected as 500 ppm except for one isolate which was highly resistant, and 500 ppm for all isolates except for two isolates which were highly sensitive respectively. Magnesium in different concentrations (500 and 2000 ppm) showed synergistic activity with erythromycin and clindamycin. Results reveal high heavy metal tolerance among antibiotic resistant MRSA strains, in addition to the presence of virulence factors. Upcoming studies must be focused on the combination of sub-inhibitory concentration of different heavy metals with the available antibiotics.
MRSA, biofilm, MLSB, heavy metals, S. aureus
Staphylococcus aureus is a bacterial pathogen, which is considered as one of the most common agents in a variety of human infections including skin, soft tissue as well as eye1. Those bacterial species shows some virulence factors which are tightly regulated specially during bacterial growth and pathogenesis process 2.
Formation of biofilms is one of the important virulence factors that provide hard bacterial growth mode making them resist the unfavorable conditions more than those without biofilms3. A central role in the regulation of such virulence is controlled by bacterial quorum-sensing system4. Bacterial biofilm formation ability always allow more advantages for bacterial cell survival compared to planktonic growth, including protection from shear stress, disinfectants, and antibiotics by its special structure5-7. This mode of bacterial growth induces cellular dormancy associated with multidrug tolerance, and non-heritable phenotype that differs from classic mechanisms of antibiotic resistance8,9.
Resistance to antimicrobial agents has become a difficult problem all over the world10,11. Nowadays, the emergence of methicillin-resistant S. aureus (MRSA) became a great concern as it causes life-threatening nosocomial and community-acquired infections12. Due to the limited number of effective antibiotics, treatment of such infections is sometimes considered very difficult13. S. aureus isolates qualify to have a resistance to erythromycin which is commonly associated with resistance to other macrolides. Three genes were found in S. aureus responsible for this resistance viz. ermA, ermB and ermC, and encode methylase enzymes which have a role in modifying the ribosomal target site leading to S. aureus MLSB phenotype14.
Treatment of patients infected with S. aureus with clindamycin harboring inducible clindamycin resistance (iMLSb phenotype) might lead to the evolution of resistant strains (cMLSB) and subsequently, causing more therapeutic failure15. Erythromycin and clindamycin are different classes of antibiotics which bind to the 50S ribosomal subunit of bacteria and inhibit protein synthesis. S. aureus resistance to these antibiotics is created by methylation of the target site on the ribosome mostly related to methylase gene erm (rRNA)16.
The widespread metal contamination of human surrounding is considered a recalcitrant selection pressure in bacteria with clinical and environmental importance that donates some antibiotic resistance factors17. Indeed, bacteria are continually changing their genetic compositions, which makes them able to adapt and alter their living conditions. Therefore, the heavy metal-induced bacterial resistance plays an important role in the development of bacteria18 and may play an important role in disseminating bacterial antibiotic resistance, but the selective effects of heavy metals on bacterial antibiotic resistance are largely unclear. To investigate this, the effects of heavy metals on antibiotic resistance were studied in a genome-sequenced bacterium, LSJC7. The results showed that the presence of arsenate, copper, and zinc were implicated in fortifying the resistance of LSJC7 towards tetracycline. The concentrations of heavy metals required to induce antibiotic resistance, i.e., the minimum heavy metal concentrations (MHCs. Much like antibiotics, heavy metals often have toxic effects on bacteria. Therefore, due to their antimicrobial properties, they are mostly incorporated into products19.
Through our investigation, we evaluated the antibiotics resistance of ten clinical S. aureus isolates, besides detection of their MLBS phenotypes. The heavy metal tolerance among antibiotic-resistant MRSA strains was investigated. In addition, the occurrence of S. aureus virulence factors such as biofilm formation ability as well as hemolytic and lipolytic activities were assessed.
Bacterial strains
Ten S. aureus isolates used in the current study were collected from El-Kasr el Ainy hospital, Cairo, Egypt. Initially, the strains of S. aureus were identified using traditional biochemical tests including Gram staining, catalase, coagulase (both slide and tube), and DNase tests, and confirmed by growing in selective Mannitol Salt Agar (MSA) medium20.
Antibiogram profiling
Antibiotic sensitivity was performed by using the standard disk diffusion method on Müller–Hinton (MH; Laboratories Conda SA, Madrid, Spain) agar plate according to recommended CLSI guidelines21. Standard antimicrobial disks (Oxoid, Basingstoke, UK) representing seven different drug classes such as amikacin (30 µg), ciprofloxacin (5 µg), amoxicillin/clavulanic (30 µg), cefoxitin (FOX), sulphamethoxazole-trimethoprim (1.25/23.75 µg), ampicillin (10 µg), erythromycin (15 µg), and clindamycin (2 µg) were used.
Detection of Macrolide-Lincosamide-Streptogramin B (MLSB) resistance phenotypes
MLSB resistance pattern was evaluated for all S. aureus isolates using the D-test13. At a distance of 15mm (edge to edge), erythromycin (15 μg) and clindamycin (2 μg) discs were placed on MH agar plate which was previously inoculated with 0.5 MacFarland bacterial suspensions. Following overnight incubation at 37°C, D shaped zone formed around clindamycin in the area between the two discs indicated positive result. The results were interpreted as erythromycin-sensitive, clindamycin-sensitive (S); erythromycin-resistant, clindamycin-resistant (constitutive MLSB); erythromycin-resistant, clindamycin-sensitive, D-test positive (inducible MLSB); erythromycin-resistant, clindamycin-sensitive, D-test negative (MS)22.
Characterization of the S. aureus enzymatic activity
Lipolytic activity was carried out using agar plates containing tween 20 (1% v/v) as described previously 23. The plates were inoculated with a single colony of a culture on nutrient agar and incubated for (1-5) days at 37°C. The turbid zone around colonies, with the change to blue color after the addition of a saturated solution of CuSO4 .5H2O reagent, indicates a positive result.
Hemolysis test on blood agar with defibrinated sheep blood 5% (v/v) was used. Isolates were inoculated and incubated for a day at 37 °C. After incubation, the formation of zones around the bacterial growth was detected and characterized as a (partial hemolysis), β (complete hemolysis), and γ (no hemolysis) depending on the activities of each bacterial strain24.
Microtiter plate biofilm assay
In order to perform the biofilm assay, wells of the 96 polystyrene plate were first filled with 100μl of the bacteria inoculated in Brain Heart Infusion broth, at which uninoculated broth was considered as negative control. After 24 hours incubation at 37°C, the contents of wells were gently removed. The wells were washed as recommended, dried, and the adherent bacteria biofilms formed were then stained with 1% aqueous solution of crystal violet (CV). After 10 minutes, excess stain was washed gently, and the plate was kept for drying. To solubilize the stained adherent biofilm, absolute ethanol was used25.
By using an auto-reader (Stat Fax-2100; GMI, Inc. USA), the optical density was measured at a wavelength of 570 nm. Each experiment was performed three times. Optical density cut-off value (O.Dc) was calculated as mentioned7.
Antibacterial effect of antibiotics against bacterial isolates
Antibacterial activity assay of antibiotics was carried out by microtiter broth dilution method26. As a diluent, MH broth was used, where about 106 CFU/mL cells (0.5 MacFarland bacterial cultures) were inoculated, at which the final volume in each microtiter plate well was 100 µl and antibiotics concentration ranged from 0.25-16 µl/ml, 0.06-4 µl/ml, and 0.125-8 µl/ml for gentamycin, ciprofloxacin, and erythromycin respectively. After 24 hours incubation at 37°C, the minimum inhibitory concentration (MIC) values, defined as the lowest antibiotic concentration which inhibited visual bacterial growth, were measured.
Detection of heavy metals/essential elements maximum tolerance concentration (MTC)
The MTC values of heavy metals/essential elements for the bacterial isolates were determined by agar dilution method, using nutrient agar supplemented with different heavy metals like: copper (Cu2+), cobalt (Co2+), zinc (Zn2+) and iron (Fe2+), and different essential elements like: potassium (K+) and magnesium (Mg2+). The concentrations of the utilized metals included: 500, 1500 and 2000 ppm. After 18 hours incubation at 37°C, the MTC, defined as the highest metal/element concentration that allow the bacterial growth, were measured27.
Effect of biofilm formation
The antibiofilm activity of different antibiotics at sub-inhibitory concentrations was done as described with minor modifications28. In case of no MIC detected, the ½ MIC represents the highest antibiotic concentration. 100 μl of bacterial suspension (0.5 McFarland) were dispensed into each well of 96-well polystyrene microtiter plates, and different antibiotic concentrations (100 μl) were added, and plates were incubated at 37°C for 18 hours. Antibiotic free wells (biofilm growth) act as positive controls. After incubation, the medium and non-adherent cells were removed and wells were washed three times with sterile PBS. The plates were well dried and then the dye was re-solubilized with absolute ethanol. The OD of each well was measured at 570 nm using ELISA reader. Each assay was performed in duplicates.
The antibiotic/ potassium (K+) and magnesium (Mg2+) synergistic assay
Synergistic activity was assessed by using the agar disk diffusion method using antibiotic disks of erythromycin (15 µg), and clindamycin (2 µg). MH agar supplemented with different elements; K+ and Mg2+ concentrations was used. The bacterial inoculum of 0.5 McFarland concentrations was applied on the surface of the agar plate and antibiotic disks were placed onto the inoculated agar surface. After incubation for 18 hours at 37°C, the zones of growth inhibition around each of the antibiotic disks were measured. MH agar plates without K+ and Mg2+ supplementation were considered as negative control6.
Statistical analysis
Chi-square test (χ2) was conducted to analyze the significance of the resistance level to various antibiotics. A value of p < 0.05 was considered to be statistically significant. The calculated mean values are presented with standard deviation (SD) bars using the Duncan test using SPSS software (Statistical Package for the Social Sciences, ver. 12.0, New York)
Antibiogram profiling
In this study, ten methicillin-resistant S. aureus bacterial isolates were used. The sensitivity test was performed using the Kirby-Bauer antibiogram method using antibiotic discs representing seven different classes of antibiotics and results were interpreted according to CLSI. Results presented in Table (1) showed that high resistance to ampicillin and amoxicillin/clavulanic by percentages of 100% was obtained. On the other hand, clindamycin was found to be the most effective antibiotic against the isolates with 70% of strains exhibiting sensitivity to this drug (Table 1).
Table (1):
Antibiotic susceptibility pattern among MRSA isolates.
| Antibiotic class | Antibiotics | Antibiogram profile | p-value* | |||
|---|---|---|---|---|---|---|
| Resistant | Non resistant | |||||
| No. | % | No. | % | |||
| Aminoglycosides | Amikacin | 5 | 50 | 5 | 50 | 0.740 |
| β-lactam | Cefoxitin | 10 | 100 | 0 | 0 | – |
| Amoxicillin/clavulanic | 10 | 100 | 0 | 0 | – | |
| Fluoroquinolones | Ciprofloxacin | 8 | 80 | 2 | 20 | 0.678 |
| Sulphonamides | Sulphamethoxazole-trimethoprim | 3 | 30 | 7 | 70 | 0.717 |
| Aminopenicillins | Ampicillin | 10 | 100 | 0 | 0 | – |
| Macrolide | Erythromycin | 9 | 90 | 1 | 10 | 0.580 |
| Lincosamide | Clindamycin | 3 | 30 | 7 | 70 | 0.717 |
*A value of p < 0.05 was considered to be statistically significant
Detection of MLSB resistance phenotypes
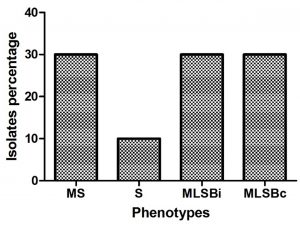
Resistant phenotypes among S. aureus strains were detected using D test. Different resistance phenotypes were detected as constitutive MLSB, inducible MLSB, S, and MS by percentages of 10%, 30%, 30% and 30% respectively (Fig. 1).
Fig. 1. Resistant phenotypes among S. aureus strains. Erythromycin-sensitive, Clindamycin-sensitive (S); Erythromycin-resistant, Clindamycin-resistant (constitutive MLSB); Erythromycin-resistant, Clindamycin-sensitive, D-test positive (inducible MLSB); Erythromycin-resistant, Clindamycin-sensitive, D-test negative (MS).
Characterization of the enzymatic activity and biofilm formation ability
The virulence factors of MRSA including lipolytic and hemolytic activity were tested among the ten strains (Table 2), the results showed that 50% of isolates had lipolytic activity, whereas 70% of them had hemolytic activity. Regarding the biofilm formation of MRSA, it was observed that all tested isolates were capable of producing biofilm and most of the isolates (70%) forms strong biofilms while 30% were categorized as moderate biofilm formers (Table 2).
Table (2):
Virulence factors and resistance phenotypes assessment among S. aureus stains.
| Strains | MLSB resistance phenotypes | Virulence factors | |||
|---|---|---|---|---|---|
| Enzymatic activity | **Biofilm formation | ||||
| *Lipolytic activity | Hemolytic activity | Average O.D. ± SD | Biofilm ability categorization | ||
| S1 | MS | + | α hemolysis | 3.469 ± 0.057 | +++ |
| S2 | MLSBi | – | γ hemolysis | 3.131 ± 0.053 | +++ |
| S3 | MLSBi | + | γ hemolysis | 3.661 ± 0.008 | +++ |
| S4 | MS | – | α hemolysis | 2.797 ± 0.192 | ++ |
| S5 | MLSBi | + | β hemolysis | 3.382 ± 0.171 | +++ |
| S6 | MS | + | β hemolysis | 3.569 ± 0.020 | +++ |
| S7 | S | + | α hemolysis | 3.312 ± 0.074 | +++ |
| S8 | MLSBc | – | γ hemolysis | 3.300 ± 0.050 | +++ |
| S9 | MLSBc | – | β hemolysis | 2.123 ± 0.404 | ++ |
| S10 | MLSBc | – | β hemolysis | 1.993 ± 0.506 | ++ |
MLSBi: inducible MLSB; MLSBc: constitutive MLSB.
*Lipolytic activity was interpreted as negative (-) or positive (+).
**Biofilm production was interpreted as: negative (-), weak (+), moderate (++) strong (+++).
Minimum inhibitory concentrations of different antibiotics
The susceptibility of three antibiotics (gentamycin, ciprofloxacin and erythromycin) was confirmed using micro-broth dilution method (Table 3). High resistance rates among tested MRSA strains were observed.
Table (3):
Minimum inhibitory concentration values of different antibiotics.
| Isolates | Minimum inhibitory concentration (mg/l) | ||
|---|---|---|---|
| Ciprofloxacin | Gentamycin | Erythromycin | |
| S1 | 1 | ≥ 16 | ≥ 8 |
| S2 | ≥ 4 | ≥ 16 | ≥ 8 |
| S3 | ≥ 4 | ≥ 16 | ≥ 8 |
| S4 | ≥ 4 | ≥ 16 | ≥ 8 |
| S5 | ≥ 4 | ≥ 16 | ≥ 8 |
| S6 | 2 | 4 | ≥ 8 |
| S7 | ≥ 4 | 8 | 2 |
| S8 | ≥ 4 | ≥ 16 | ≥ 8 |
| S9 | ≥ 4 | ≥ 16 | ≥ 8 |
| S10 | ≥ 4 | 16 | ≥ 8 |
Detection of heavy metals maximum tolerance concentration (MTC)
Detection of heavy metals MTC was performed using MH supplemented with three concentrations (500, 1500 and 2000 ppm) of different heavy metals used. The recorded results in Table 4 demonstrated that the bacteria can grow at high concentrations of K+ and Mg2+. On the other hand, the MTC of heavy metals was evaluated as 500 ppm for all isolates in case of cobalt and iron. For zinc, the MTC was 500 ppm in all isolates except for isolate number 7 which was highly resistant, while in case of copper the MTC was 500 ppm for all isolates except for isolates 2 and 5 which were highly sensitive even in the lowest concentration.
Table (4):
Maximum tolerance concentration of different heavy metals/elements by agar dilution method.
| Strains | Used concentrations (ppm) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Essential elements | Heavy metals | |||||||||||||||||
| K | Mg | Cu | Co | Zn | Fe | |||||||||||||
| 500 | 1500 | 2000 | 500 | 1500 | 2000 | 500 | 1500 | 2000 | 500 | 1500 | 2000 | 500 | 1500 | 2000 | 500 | 1500 | 2000 | |
| S1 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S2 | + | + | + | + | + | + | – | – | – | + | – | – | + | – | – | + | – | – |
| S3 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S4 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S5 | + | + | + | + | + | + | – | – | – | + | – | – | + | – | – | + | – | – |
| S6 | + | + | + | + | + | + | + | – | – | + | – | – | + | + | + | + | – | – |
| S7 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S8 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S9 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
| S10 | + | + | + | + | + | + | + | – | – | + | – | – | + | – | – | + | – | – |
K: potassium; Mg: magnesium; Cu: copper; Co: cobalt; Zn: zinc; Fe: iron; (+): Bacterial growth; (-): No bacterial growth
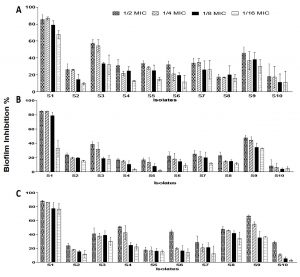
Fig. 2. Effect of different antibiotics concentrations on MRSA strains biofilm formation. (a) Gentamycin (b) Ciprofloxacin and (c) Erythromycin. The bars on the graph represent mean ± SD as a percentage of biofilm inhibition/eradication of triplicate independent experiments (n = 2).
Antibiofilm activity of commercial antibiotics
The effect of three antibiotics was studied to inhibit the MRSA strains biofilm formation ability. Different inhibition percentages were showed (Fig. 2). Generally, low biofilm inhibition percentages were observed for the three tested antibiotics, at which the best inhibition percentages observed by using ½ MIC antibiotics concentration, followed by ¼, 1/8 and 1/16 MICs. The most sensitive isolates which showed reduction in biofilm formation ability were S1, S3 and S9.
Antibiotic/heavy metals synergistic assay
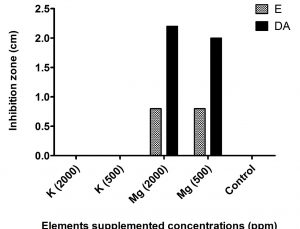
Synergistic activity of K+ and Mg2+ at concentrations of 500 and 2000 ppm (Fig. 3), showed that enhanced inhibition zones were detected in case of magnesium supplementation, while on the other hand no zones were detected in case of potassium supplementation.
Recently, Staphylococcus aureus has drawn much attention among researchers due to its adaptation ability to a range of antibiotics causing resistance29, and these bacterial characters can complicate infection treatment and control, specially MRSA strains30. Over the past few years, reports about MRSA prevalence were obtained showing different prevalence over countries, at which MRSA prevalence of 53.4% in East Africa 29, 29% in United Arab Emirates (UAE)31, 34.2% in India 32.
In the present investigation, the antibiotic of S. aureus reported a high resistance to ampicillin and amoxicillin/clavulanic by percentages of 100%, followed by resistance to erythromycin (90%) and ciprofloxacin (60%). The resulted ciprofloxacin resistance percentage is in agreement with many reported studies, However, noticeable elevated rate of erythromycin resistance was recorded compared to other reports12,33,34. On the other hand, the bacteria showed high sensitivity to clindamycin, which is in line with the study reporting 73.2% sensitivity percentage33, but considered higher than the percentage reported in Palestine (34.8%)34.
Resistance to macrolides, lincosamides, and group B streptogramins is commonly referred to as “MLSB resistance”. The erm gene encodes methylation of the 23S rRNA–binding site that is shared by these three drug classes10,12. Erythromycin and clindamycin are antimicrobials which belong to the MLSB group widely recommended to treat staphylococcal infections 35. In this study, different MLSB resistance phenotypes percentages for S. aureus including S, cMLSB, iMLSB and MS respectively were recorded. In literature survey of many countries, it was observed that in India, 64.8% had cMLSB, 25% had iMLSB and 8% had the MS phenotype among the tested MRSA isolates15. In South Ethiopia, inducible clindamycin resistance was detected in 19/22 of the erythromycin-resistant strains36. The prevalence of iMLSB and cMLSB phenotypes was 11.48% and 29.25% respectively, where both phenotypes predominated in MRSA strains were reported37. This reported resistance to macrolides is suggested to be due to modification of the bacterial intracellular drug-binding site on the ribosome10.
Indeed, several virulence factors involved in the pathogenesis character of S. aureus strains have been described in the literature38. Because of the bacterial ability to grow in biofilms, this bacterial pathogens exhibited drug tolerance to a broad-spectrum of antibiotics, which is attributed to the biofilm itself. Pathogens are able to produce biofilms, which can be explained by many factors including the decreased permeability of the drug inside the bacterial cells, the appearance of persisting bacteria, and intracellular bacterial survival39. In literature, different reports for biofilm occurrence were found, Piechota et al., (2018) reported that strong biofilm was formed by 39.7% of MRSA40. A percentage of 50% was demonstrated in another study as weak biofilm producers followed by moderate and strong biofilm ones by 27% and 23% respectively41. Furthermore, higher biofilm formation was also reported at different percentages of 69.8%, 68.3%, and 53.5%1,5,42.
Low inhibition percentages were observed by using different antibiotics concentrations (Fig. 3). Recently, sub-inhibitory concentrations of different commercially used antibiotics were tested on the Staphylococcus aureus biofilms, and as a result different reductions in those biofilms were observed26,43,44. Antibiotics at sub-MIC levels were found to affect some biofilm virulence expression, suggesting that it is antibiotic dose dependent28,45.
Potentially toxic metal contaminants may interact with ecosystem native inhabitant; therefore those bacteria can develop some mechanisms of tolerance allowing them to survive46. This co-occurrence of such antibiotic/metal resistance has been reported 47. In this study, high MTC values were recorded, 500 ppm for all isolates, in case of cobalt, iron and zinc (except for isolate 7) in addition to copper (except for isolates 2 and 5). On the other hand, the bacteria were grown normally at elevated concentrations of K+ and Mg2+.
The first-row d-block metal ions, which are manganese (Mn), iron (Fe), cobalt (Co), nickel (Ni), copper (Cu), and zinc (Zn), are essential micronutrients. Cu and Zn are now recognized to be involved in the phagosomal killing of bacteria engulfed by macrophages, defined as an important defense mechanism48. The rapid killing of bacteria is thought to be due to cellular damage caused by very high concentrations of Cu dissolving from the surface, which causes membrane disturbance and rupture, coupled with internal oxidative burst due to the generation of ROS leading to further cellular destruction, including degradation of plasmid and chromosomal DNA49.
According to our results, no effect for both K+ and Mg2+ was detected even in the highest concentration. On the other hand, by using predictable synergistic disc diffusion method, Mg2+ supplementation at concentrations of 500 and 2000 ppm showed enhancement activity for both antibiotics used (Fig. 2). K+ is responsible for many cellular processes, including regulation of intracellular pH, maintenance of turgor, and activation of enzymes50. Whereas, Mg2+ is well known to be important in the normal homeostatic mechanisms of the cells51,52.
Our results reveal high heavy metal tolerance among antibiotic-resistant MRSA strains, in addition to the presence of virulence factors such as biofilm formation and enzymatic activity (hemolytic and lipolytic). The supplementation of Mg at different concentrations (500 and 2000 ppm) showed synergistic activity with the antibiotics; erythromycin and clindamycin. To enhance the antibiotic activities, upcoming studies must be focused on the combination of a sub-inhibitory concentration of different heavy metals with the available antibiotics.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Neopane P, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25-32.
Crossref - Jafari-Sales A, Farhadi F, Ezdiyadi M, Tarbiat-Nazloo D. Study of antibiotic resistance pattern in methicillin-resistant Staphylococcus aureus isolated from clinical samples of hospitals in Tabriz – Iran. Int J Biomed Public Heal. 2018;1(2):71-75.
- Metwally M, Ali S, Khatab I, El-Sayed M. Antibacterial Potential of some Seaweeds Species to Combat Biofilm-producing Multi-drug Resistant Staphylococcus aureus of Nile Tilapia. Egypt J Bot. 2020;60(1):9-24.
Crossref - Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. Expression of Virulence Factors by Staphylococcus aureus Grown in Serum. Appl Environ Microbiol. 2011;77(22):8097-8105.
Crossref - Haddad O, Merghni A, Elargoubi A, Rhim H, Kadri Y, Mastouri M. Comparative study of virulence factors among methicillin resistant Staphylococcus aureus clinical isolates. BMC Infect Dis. 2018;18:560.
Crossref - Mohamed SH, Khalil MS, Azmy M. In vitro Efficiency of Ampicillin, Thymol and Their Combinations against Virulence Strains of Klebsiella pneumoniae. Int J Pharm Res. 2019;11(3):315-321.
- Mohamed SH, Khalil MS, Mohamed MSM, Mabrouk MI. Prevalence of antibiotic resistance and biofilm formation in Klebsiella pneumoniae carrying fimbrial genes in Egypt. Res J Pharm Technol. 2020;13(7):3051-3058.
Crossref - Watters CM, Burton T, Kirui DK, Millenbaugh NJ. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect Drug Resist. 2016;9:71-78.
Crossref - Mohamed SH, Mohamed MSM, Khalil MS, Mohamed WS, Mabrouk MI. Antibiofilm activity of papain enzyme against pathogenic Klebsiella pneumoniae. J Appl Pharm Sci. 2018;8(06):163-168.
Crossref - Ii JSL, Jorgensen JH. Inducible Clindamycin Resistance in Staphylococci : Should Clinicians and Microbiologists be Concerned ? Antimicrob Resist. 2005;40(2):280-285.
Crossref - Mohamed SH, Khalil MS. DNA Sequencing of bla SHV Genes among Uropathogenic Klebsiella pneumoniae Harboring bla CTX-M-15. Int J Pharm Res. 2020;12:443-449.
Crossref - Ahmed MO, Abuzweda AR, Alghazali MH, et al. Misidentification of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in Tripoli, Libya. Libyan J Med. 2010;5:5230.
Crossref - Navidinia M. Detection of inducible clindamycin resistance (MLSBi) among methicillin- resistant Staphylococcus aureus (MRSA) isolated from health care providers. J Paramed Sci. 2015;6(1):91-96.
- Kareem SM, Jubori SSA-, Ali M. Prevalence of erm Genes among Methicillin Resistant Staphylococcus aureus MRSA Iraqi Isolates. Int J Curr Microbiol App Sci. 2015;4(5):575-585.
- Singh T, Deshmukh AB, Chitnis V, Bajpai T. Inducible clindamycin resistance among the clinical isolates of Staphylococcus aureus in a tertiary care hospital. Int J Heal Allied Sci. 2016;5(2):111-114.
Crossref - Talebi G, Hashemia A, Goudarzi H, et al. Survey of ermA, ermB, ermC and mecA genes among Staphylococcus aureus isolates isolated from patients admitted to hospitals in Tehran, Iran by PCR and sequencing. Biomed Res. 2019;30(2):259-263.
Crossref - Baker-austin C, Wright MS, Stepanauskas R, Mcarthur JV. Co-selection of antibiotic and metal resistance. TRENDS Microbiol. 2006;14(4):176-182.
Crossref - Chen S, Li X, Sun G, Zhang Y, Su J, Ye J. Heavy Metal Induced Antibiotic Resistance in Bacterium LSJC7. Int J Mol Sci. 2015;16(10):23390-23404.
Crossref - Eggers S, Safdar N, Malecki KMC. Heavy metal exposure and nasal Staphylococcus aureus colonization : analysis of the National Health and Nutrition Examination Survey (NHANES). Environ Heal. 2018;17(2):1-10.
Crossref - Mohamed MSM, Abdallah AA, Mahran MH, Shalaby M. Potential Alternative Treatment of Ocular Bacterial Infections by Oil Derived from Syzygium aromaticum Flower (Clove). Curr Eye Res. 2018;43(7):873-881.
Crossref - The Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100-S26. 2016.
- Dash M. Detection of inducible and constitutive clindamycin resistance among Staphylococcus aureus isolates in a tertiary care hospital, Eastern India. Avicenna J Med. 2016;6(3):75-80.
Crossref - Mohammad HH. Phenotypic Investigation for Virulence factors of Pyocine producing Pseudomonas aeruginosa Isolated from Burn Wounds, Iraq. Int J Sci Eng Res. 2013;4(7):2114-2121.
- Hasan R, Acharjee M, Noor R. Prevalence of vancomycin resistant Staphylococcus aureus (VRSA) in methicillin resistant S. aureus (MRSA) strains isolated from burn wound infections. Tzu Chi Med J. 2016;28(2):49-53.
Crossref - Mohamed SH, Salem D, Azmy M, Fam NS. Antibacterial and antibiofilm activity of cinnamaldehyde against carbapenem-resistant Acinetobacter baumannii in Egypt : In vitro study. J Appl Pharm Sci. 2018;8(11):151-156.
Crossref - Mohamed M, Mostafa H, Mohamed S, El-Moez SA, Kamel Z. Combination of Silver Nanoparticles and Vancomycin to Overcome Antibiotic Resistance in Planktonic/Biofilm Cell from Clinical and Animal Source. Microb Drug Resist. 2020.
Crossref - Afzal AM, Rasool MH, Waseem M, Aslam B. Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express. 2017;7(184):184.
Crossref - Mohamed SH, Mohamed MSM, Khalil M, Azmy M, Mabrouk M. Combination of essential oil and ciprofloxacin to inhibit/eradicate biofilms in multidrug-resistant Klebsiella pneumoniae. J Appl Microbiol. 2018;125(1):84-95.
Crossref - Wangai FK, Masika MM, Maritim MC, Seaton RA. Methicillin-resistant Staphylococcus aureus (MRSA) in East Africa: red alert or red herring? BMC Infect Dis. 2019;19:596.
Crossref - Kateete DP, Bwanga F, Seni J, et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob Resist Infect Control. 2019;8:94.
Crossref - Jalaf M Al, Fadali H, Alanee R, et al. Methicillin resistant Staphylococcus aureus in emergency department patients in the United Arab Emirates. BMC Emerg Med. 2018;18:12.
Crossref - Perala BMK, Koripella RL, Vani TMSS, Lakshmi N. Prevalence of Methicillin Resistant Staphylococcus aureus in A Tertiary Care Hospital. J Dent Med Sci. 2016;15(5):29-31.
Crossref - Abdullahi N, Iregbu KC. Methicillin-Resistant Staphylococcus aureus in a Central Nigeria Tertiary Hospital. Ann Trop Pathol. 2018;9(1):6-10.
Crossref - Hadyeh E, Azmi K, Seir RA, Abdellatief I, Abdeen Z. Molecular Characterization of Methicillin Resistant Staphylococcus aureus in West Bank-Palestine. Front Public Heal. 2019;7:130.
Crossref - Martins MA, Santos S de LV dos, Leao LSN de O, Araujo NP, Bachion MM. Prevalence of resistance phenotypes in Staphylococcus aureus and coagulase-negative isolates of venous ulcers of primary healthcare patients. Rev Soc Bras Med Trop. 2012;45(6):717-722.
Crossref - Mama M, Aklilu A, Misgna K, Tadesse M, Alemayehu E. Methicillin-and Inducible Clindamycin-Resistant Staphylococcus aureus among Patients with Wound Infection Attending Arba Minch Hospital, South Ethiopia. Int J Microbiol. 2019;2019:2965490.
Crossref - Adhikari RP, Shrestha S, Barakoti A, Amatya R. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infect Dis. 2017;17:483.
Crossref - Bien J, Sokolova O, Bozko P. Characterization of Virulence Factors of Staphylococcus aureus : Novel Function of Known Virulence Factors That Are Implicated in Activation of Airway Epithelial Proinflammatory Response. J ofPathogens. 2011;2011:601905.
Crossref - Mohamed SH, Elshahed MMS, Saied YM. Evaluation of Honey as an antibacterial agent against drug-resistant uropathogenic E. coli strains. Res J Pharm Tech. 2020;13(8):3720-3724.
- Piechota MB, Kot B, Frankowska-maciejewska A, Gru A, Wo A. Biofilm Formation by Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Strains from Hospitalized Patients in Poland. Biomed Res Int. 2018;2018:4657396.
Crossref - Sohail M, Latif Z. Molecular analysis, biofilm formation, and susceptibility of methicillin-resistant Staphylococcus aureus strains causing community- and health care-associated infections in central venous catheters. Rev Soc Bras Med Trop. 2018;51(5):603-609.
Crossref - Jeong-ok C, Jae I, Jung S, et al. Investigation of Biofilm Formation and its Association with the Molecular and Clinical Characteristics of Methicillin-resistant Staphylococcus aureus. Osong Public Heal Res Perspect. 2013;4(5):225-232.
Crossref - Yang B, Lei Z, Zhao Y, et al. Combination Susceptibility Testing of Common Antimicrobials in Vitro and the Effects of Sub-MIC of Antimicrobials on Staphylococcus aureus Biofilm Formation. Front Microbiol. 2017;8:2125.
Crossref - Lazaro-Diez M, Remuzgo-Martinez S, Rodriguez-Mirones C, et al. Effects of Subinhibitory Concentrations of Ceftaroline on Methicillin-Resistant Staphylococcus aureus (MRSA) Biofilms. PLoS One. 2016;11(1):e0147569.
Crossref - Haddadin RNS, Saleh S, Al-Adham ISI, Buultjens TEJ, Collier PJ. The effect of subminimal inhibitory concentrations of antibiotics on virulence factors expressed by Staphylococcus aureus biofilms. J Appl Microbiol. 2010;108(4):1281-1291.
Crossref - Mohamed MS, El-Arabi NI, El-Hussein A, El-Maaty SA, Abdelhadi AA. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol.(Praha). 2020;65(4):687-696:
Crossref - Yazdankhah S, Skjerve E, Wasteson Y. Antimicrobial resistance due to the content of potentially toxic metals in soil and fertilizing products. Microb Ecol Health Dis. 2018;29:1548248.
Crossref - Djoko KY, Ong CY, Walker MJ, Mcewan AG. The Role of Copper and Zinc Toxicity in Innate Immune Defense against Bacterial Pathogens. J Biol Chem. 2015;290(31):18954-18961.
Crossref - Grass G, Rensing C, Solioz M. Metallic Copper as an Antimicrobial Surface. Appl Environ Microbiol. 2011;77(5):1541-1547.
Crossref - Schramke H, Laermann V, Tegetmeyer HE, Brachmann A, Jung K, Altendorf K. Revisiting regulation of potassium homeostasis in Escherichia coli: the connection to phosphate limitation. Microbiology Open. 2017;6(3):e438.
Crossref - Saris NL, Mervaala E, Karppanen H, Khawaja JA, Lewenstam A. Magnesium An update on physiological, clinical and analytical aspects. Clin Chim Acta. 2000;294(1):1-26.
Crossref - Staiger MP, Pietak AM, Huadmai J, Dias G. Magnesium and its alloys as orthopedic biomaterials : A review. Biomaterials. 2006;27(9):1728-1734.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.