ISSN: 0973-7510
E-ISSN: 2581-690X
Pathogenic micro-organisms from contaminated food are capable of causing serious infections and hence this issue has become a healthcare problem globally. The contamination may occur, either directly by an infected food handler, or indirectly through contact with a food contact surface that has been previously contaminated by an infected food handler. The current study was aimed to detect the pathogenic bacteria from food handlers in Ha’il region of Saudi Arabia. In this study, 152 bacterial isolates were collected from 50 food handlers. Identification of bacterial isolates was performed by conventional methods as well as by automated methods using Microscan, VITEK 2 and MALDI-TOF-MS. The results of conventional methods showed, 28.3% (43/152) bacterial isolates were Gram-positive and 71.7% (109/152) were Gram-negative. Among the Gram-positive isolates, E. faecalis, S. aureus and E. faecium were found to be 8.5% (13/152), 7.2% (11/152) and 4% (6/152) respectively. Among Gram-negative isolates, P. mirabilis, E. coli, E. cloacae and K. pneumoniae were found to be 12.5% (19/152), 11% (17/152), 11% (17/152) and 10.5% (16/152) respectively. The antibiotic susceptibility of the bacterial isolates in our study showed that 100% S. aureus were ciprofloxacin resistant. Additionally, 62% E. faecalis were resistant to gentamicin, 19% E. coli and 12% K. pneumoniae were found to be ESBL positive. The identification of bacterial isolates by 3 automated methods, showed that 93% (141/152), 94% (143/152) and 96% (146/152) bacterial isolates were correctly identified by Microscan, VITEK 2 and MALDI-TOF-MS respectively. Thus MALD-TOF-MS proves to be the economical, fast and accurate method for identification of food borne pathogens. Incorporating this technique into food microbiology would lead to more successful and rapid identification of pathogenic bacteria from food sources.
Food contamination, Pathogenic bacteria, MALDI-TOF-MS
In the food processing industry, a food handler’s role is one of the most important in ensuring the safety of food. Food handler is an essential part in the chain of preparation, cooking, packaging and delivery of food. A food handler is directly involved in, packaging or unpackaging food, food equipment and utensils, or food contact surfaces1. In order to make sure that the food is safe and free from any contamination, a food handler must fulfill the requirements to ensure the food hygiene. During the preparation, processing, delivery and serving of food, a food handler is capable of being a potential source of bacteria that causes foodborne diseases by introducing these pathogens in to the food. It has been found that incorrect practices in the food industry by a food handler are responsible for about 97% of foodborne ailments2-3.
The contamination of food with harmful micro-organisms may occur, either directly by an infected food handler, or indirectly through contact with a food contact surface that has been previously contaminated by an infected food handler4. In addition to pathogens, toxins, and other contaminants of the food also pose a serious threat to human health, and lead to high morbidity and mortality. Among many micro-organisms, some of the various bacterial pathogens that have been found to be the frequent contaminants of food are Salmonella, Listeria, S. aureus, Campylobacter, Trichinosis, E. coli, Campylobacter and Clostridium. These microbes cause severe infections with high morbidity, and majority of these infections have been attributed to food borne transmission5. Recently, Australian institute of food safety reported that among many microbes, Salmonella, Listeria, S. aureus, Campylobacter, Trichinosis, E. coli, Campylobacter and Clostridium are the top 7 causes of food poisoning. The health department of Australia estimates that food poisoning affects around 5.4 million Australians each year6. Food borne or waterborne microbial pathogens are considered as leading causes of infections and deaths in developing countries, killing an estimated 1.9 million people annually at the global level. Even in developed countries, an estimated one-third of the populations are affected by microbiological food borne diseases each year7. The food borne infection usually involves the intestinal enteropathogenic bacteria and their transmission is helped directly or indirectly by objects contaminated with feces.
Food handlers capable of harboring and excreting enteropathogenic bacteria may contaminate foods from their feces through their fingers, then to food processing, and finally to healthy individuals8. It has been reported that the area of hand beneath fingernails works as a vector for transmission of harmful microorganisms through cross contamination as compared to other parts of the hand9. One of the major illness or infection due to bacterial contaminated food is diarrheal disease, and globally, diarrheal diseases are second only to respiratory diseases as a cause of adult death and are the leading cause of childhood death. In some parts of the world, they are responsible for more years of potential life lost than all other causes combined10. In addition to cause the food borne illness, the bacterial strains such as, Salmonella spp. and E. coli have tendency to evolve in order to exploit novel opportunities, for example fresh produce, and even generate new public health challenges like antimicrobial resistance11-13. The spread of foodborne disease due to pathogens which are highly resistant to antibiotics has become a health care issue worldwide. Additionally, the toxins produced by the bacterial strains in to the food cause a substantial loss to the food industry because a large amount of money has to be spent on analyzing and identifying preventive measures.
Currently, the gold standard; traditional culture-based methods are used to identify the majority of food-associated bacteria in the daily routine of food microbiology laboratories globally. Complete identification is a time consuming process and requires at least two days, or more for fastidious organisms. By using these phenotypic methods, sometimes, bacterial isolates with different taxonomic background and similar physiological characteristics pose a challenge and may give non reliable result.
Thus, the development of a rapid, sensitive, specific, and cost-effective analytical method is of great importance for detection of microbial contaminants in the food. Recently, many technological improvements to methods for the identification of micro-organisms, such as MALDI–TOF-MS, have successfully been incorporated in clinical microbiology laboratories globally. MALDI–TOF-MS is a useful, fast, reliable and simple technique for the correct identification of micro-organisms and several studies have highlighted the advantages and performance of MALDI–TOF-MS including, rapidity, low sample volume requirements and low reagent costs14. MALDI-TOF-MS provides a suitable platform for quick, flexible, and reliable identification of food associated microbes because of the simple protocol and shortened analysis time15. Therefore, the aim of this study was to detect the colonized pathogenic bacteria from food handlers in Ha’il region of Saudi Arabia and to compare the results using conventional methods, MALDI-TOF-MS, Microscan and Vitek 2.
Study design
In this study, a total of 50 food handlers (subjects working on meat shops) from the Ha’il region of Saudi Arabia were screened for the presence of pathogenic bacterial strains. A single non repetitive, hand swab, nasal swab and swab from any wound site were collected from each individual for screening.
Bacterial identification By conventional methods
Identification of bacterial isolates was performed by, simple staining, Gram-staining, morphology and biochemical tests.
Identification of microbes by automation methods By MALDI-TOF-MS
The identification of the microbes by MALDI-TOF-MS was performed on Bruker Daltonics instrument16, according to the manufacturer’s guidelines. In this method, a fresh colony material was smeared on a polished steel target plate (Bruker Daltonics) using a toothpick, overlaid with 1 µl of a saturated a-cyano-4-hydroxy-cinnamic acid (HCCA) matrix solution in 50% acetonitrile-2.5% trifluoroacetic acid (Bruker Daltonics), and air dried at room temperature. For the direct transfer-formic acid method, 1 µl of 70% formic acid was added to the bacterial spot and allowed to air dry before the matrix solution was added. The acquisition and analysis of mass spectra were performed by a Microflex LT mass spectrometer (Bruker Daltonics) using the MALDI Biotyper software package (version 3.0). The Bruker bacterial test standard (Bruker Daltonics) was used for calibration according to the instructions of the manufacturer. For each strain, two preparations of colony/sample material were analyzed. Standard Bruker interpretative criteria were applied to compare the data obtained with reference data base. Briefly, scores of e2.0 were accepted for species assignment and scores of e1.7 but <2.0 for identification to the genus level. Scores below 1.7 were considered unreliable.
Identification and antibiotic susceptibility by Microscan
Microscan walkaway (Siemens Healthcare Diagnostics, Sacramento, CA, USA) is an automated system used for bacterial identification and antibiotic susceptibility test. A small portion of a well isolated colony was taken and added to a Gram-positive or a Gram negative Microscan combo panel. The panel was loaded into the Microscan walkaway machine according to the manufacturer’s protocol. Results were available after 24- 48 hrs.
Identification and antibiotic susceptibility by VITEK 2
VITEK 2 (Biomerieux, France) is an automated system used for bacterial identification and antibiotic susceptibility test. A small portion of a well isolated colony was taken and added to a Gram-positive or a Gram negative Microscan combo panel. The panel was loaded into the VITEK 2 machine according to the manufacturer’s protocol. Results were available after 24- 48 hrs.
In this study, 152 bacterial isolates were collected from 50 food handlers in Ha’il region of Saudi Arabia as shown in Table 1. The results of the gold standard conventional methods showed, 28.3% (43/152) bacterial isolates were Gram-positive and 71.7% (109/152) were Gram-negative. Among the Gram-positive isolates, E. faecalis, S. aureus and E. faecium were found to be 8.5% (13/152), 7.2% (11/152) and 4% (6/152) respectively. Among Gram-negative isolates, P. mirabilis, E. coli, E. cloacae and K. pneumoniae were found to be 12.5% (19/152), 11% (17/152), 11% (17/152) and 10.5% (16/152) respectively.
Table (1):
Identification of bacterial isolates collected from food handlers in Ha’il region of Saudi Arabia using Microscan, VITEK 2 and MALDI-TOF-MS.
| Sample | Bacterial strain | No. of isolates | Correctly identified by | ||
|---|---|---|---|---|---|
| Microscan | VITEK 2 | MALDI-TOF-MS | |||
| Gram-positive | Staphylococcus sciuri | 1 | 1 | 1 | 1 |
| Staphylococcus intermedius | 1 | 1 | 1 | 1 | |
| Staphylococcus hominis | 1 | 1 | 1 | 1 | |
| Staphylococcus epidermidis | 4 | 4 | 3 | 4 | |
| Staphylococcus cohnii | 2 | 1 | 1 | 2 | |
| Staphylococcus auricularis | 2 | 2 | 1 | 1 | |
| Staphylococcus aureus | 11 | 11 | 11 | 11 | |
| Enterococcus gallinarum | 2 | 0 | 1 | 1 | |
| Enterococcus faecium | 6 | 5 | 6 | 6 | |
| Enterococcus faecalis | 13 | 13 | 12 | 13 | |
| Gram-negative | Propionibacterium | 5 | 3 | 2 | 3 |
| Proteus mirabilis | 19 | 17 | 19 | 19 | |
| Pseudomonas aeruginosa | 4 | 4 | 4 | 4 | |
| Klebsiella pneumoniae | 16 | 15 | 16 | 16 | |
| Enterobacter cloacae | 17 | 17 | 17 | 17 | |
| Enterobacter agglomerans | 11 | 10 | 10 | 10 | |
| Enterobacter aerogenes | 6 | 6 | 6 | 5 | |
| Escherichia coli | 17 | 17 | 17 | 17 | |
| Citrobacter freundii | 7 | 6 | 7 | 7 | |
| Acinetobacter lwoffii | 1 | 1 | 1 | 1 | |
| Acinetobacter baumannii | 6 | 6 | 6 | 6 | |
| Total correct identification | 152 | 141 | 143 | 146 | |
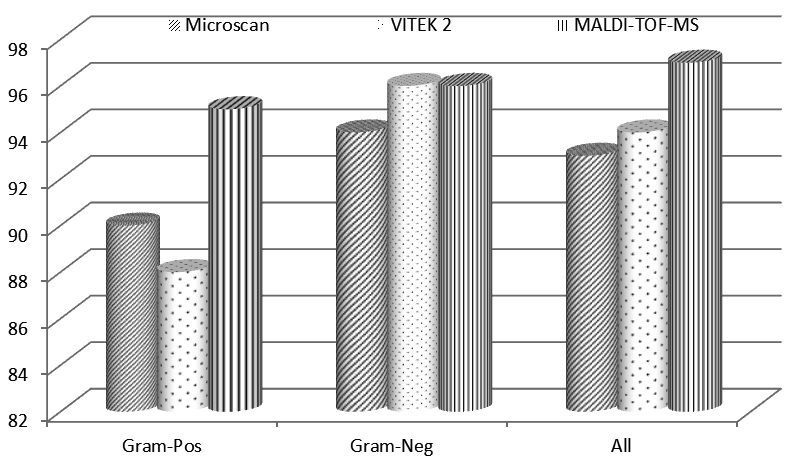
The identification of bacterial isolates was also performed by 3 automated methods, namely, Microscan, VITEK 2 and MALDI-TOF-MS. The results of identification by these automated systems showed that 93% (141/152), 94% (143/152) and 96% (146/152) bacterial isolates were correctly identified by Microscan, VITEK 2 and MALDI-TOF-MS respectively as presented in Table 1.
The comparative identification analysis of Microscan, VITEK 2 and MALDI-TOF-MS are shown in Figure 1. The data revealed that among Gram-negative isolates, MALDI-TOF-MS and VITEK 2 identified 96% isolates correctly, while as, Microscan could identify 94% isolates correctly. In the case of Gram-positive isolates, MALDI-TOF-MS identified 95% isolates correctly, while as, Microscan and VITEK 2 identified 90% and 88% isolates respectively.
The antibiotic susceptibility results showed that among Gram-positive isolates, 100% (11/11) S. aureus isolates were resistant to ciprofloxacin and 62% E. faecalis isolates were resistant to gentamicin. Among Gram-negative isolates, 19% and 12% K. pneumoniae and E. coli isolates were found to be ESBL positive.
There are many factors responsible for the contamination of food. The findings of our study indicate that food handlers i, e the subjects working on meat shops may play a vital role in transmission of pathogenic bacteria to healthy people via contaminated food. In this study, 50 food handlers were screened and 152 different bacterial strains were isolated. Among these isolates, E. faecalis, S. aureus, E. faecium, P. mirabilis, E. cloacae, E. coli and K. pneumoniae were found to be 8.5%, 7.2%, 4%, 12.5%, 11%, 11% , and 10.5% respectively . The results of our study were in agreement with a study from Iran17. In another study from Sudan conducted by Humodi et al. S. aureus was found to be the most common pathogen isolated from food handlers18. The result of current study also highlighted the significant presence of S. aureus in food handlers from Ha’il region of Saudi Arabia. The quick and reliable identification of pathogenic bacteria from the food or food handlers is essential in order to control the infections caused by these pathogens. Conventional methods of identification are time consuming and laborious, but are still considered to be the gold standard19. However, the automated systems have their own advantage and have been successfully used for identification of food borne pathogens with high sensitivity and specificity20. In our study, 3 automation methods used for identification of pathogenic bacteria from food handlers were Microscan, VITEK 2 and MALDI-TOF-MS. The results from MALDI-TOF-MS were the most accurate compared to Microscan and VITEK 2. The quick and accurate identification of pathogenic bacteria from food source is essential as many studies have shown that several antibiotic resistant bacteria have been isolated from food sources. The antibiotic susceptibility of the bacterial isolates in our study showed that all 100% S. aureus were ciprofloxacin resistant. Additionally, 62% E. faecalis were resistant to gentamicin, 19% E. coli and 12% K. pneumoniae were found to be ESBL positive.
This study reveals a high percentage of pathogenic bacteria with quite a few of these resistant to antibiotics isolated from food handlers in Ha’il region of Saudi Arabia. Furthermore, among the automated systems, MALDI-TOF-MS gave the maximum accuracy in identification of the pathogenic bacteria in this study. Thus in order to use a simple, accurate and reliable method for identification of food borne pathogens, MALDI-TOF-MS should be given a priority.
ACKNOWLEDGMENTS
This study was funded by a research grant from Ha’il University” into the article titled “Nosocomial pathogens-a single center study in Saudi Arabia.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Bertin, C.H., Rezende, M.A., Sigulem, D.M., and Morais, T.B. Hurdles at work: perceptions of hospital food handlers. Hum Resour Health, 2009; 24: 7:63.
- Lambrechts, A.A., Human, I.S., Doughari, J.H., and Lues, J.F.R. Bacterial contamination of the hands of food handlers as indicator of hand washing efficacy in some convenient food industries in South Africa. Pak J Med Sci, 2014; 30(4): 755–758.
- Howes, M., McEwen, S., Griffiths, M., and Harris, L. Food handler certification by home study: Measuring changes in knowledge and behaviour. Dairy Food Environ San, 1996; 16: 737–744.
- Alum., Eucharia Akanele., Urom., Scholastica Mgbo Otu Chukwu., Ben and Chukwu Mary Ahudie. Microbiological Contamination Of Food: The Mechanisms, Impacts And Prevention. International Journal of Scientific & Technology Research Volume 5, Issue 03, March 2016.
- Stephen Inbaraj, B., and Chen, B.H. Nanomaterial-based sensors for detection of foodborne bacterial pathogens and toxins as well as pork adulteration in meat products. Journal of Food and Drug Analysis, 2016; 24(1): 15-28.
- SALLY SANTACRUZ. Top 7 Causes of Food Poisoning. Australian Institute of Food Safety. June 2016.
- Schlundt, J., Toyofuku H., Jansen, J., and Herbst, S.A. Emerging food-borne zoonoses.Rev Sci Tech, 2004; 23: 513–515.
- WHO- Prevention and control of intestinal parasitic infections. World Health Organization, Geneva .1987; pp. 7–18. (Technical report series no .749).
- Bas, M., Ersun, A.S., and Kivanc, G. The evaluation of food hygiene knowledge, attitudes and practices of food handlers in food business in Turkey. Food Control, 2006; 17: 317–322.
- Lopez, A.D., and Mathers, C.D. Measuring the global burden of disease and epidemiological transitions: 2002-2030. Ann Trop Med Parasitol, 2006; 100(5-6): 481-499.
- Capita, R., and Alonso-Calleja, C. Antibiotic-resistant bacteria: a challenge for the food industry. Crit Rev Food Sci Nutr, 2013; 53(1): 11-48.
- Liu, Y.Y., Wang, Y., Walsh, T.R., Yi, L,X., Zhang, R., Spencer, J., et. al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infect Dis, 2016; 16(2): 161-168.
- Kirbis, A., and Krizman, M. Spread of antibiotic resistant bacteria from food of animal origin to humans and vice versa. Procedia Food Science, 2015; 5: 148 – 151.
- Al-Mogbel., M.S. Matrix assisted laser desorption/ionization time of flight mass spectrometry for identification of clostridium species isolated from Saudi Arabia. Braz. J. Microbiol, 2016; 47: 410-413.
- Pavlovic, M., Huber, I., Konrad, R., and Busch, U. Application of MALDITOF MS for the identification of food borne bacteria. Open Microbiol 2013; 7:135–141.
- Ayman Elbehiry., Eman Marzouk., Mohamed Hamada., Musaad Al-Dubaib., Essam Alyamani., et. el. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiologica, 2017; 40(4): 269-278.
- Mohtaram Nasrolaheia., Siavash Mirshafieeb., Soudeh Kholdia., Maryam Salehiana., and Masoumeh Nasrolahei. Bacterial assessment of food handlers in Sari City, Mazandaran Province, north of Iran. Journal of Infection and Public Health, 2017; 10: 171—176.
- Humodi, A.S., and Hatim, H.H. Bacteriological and parasitologi-cal assessment of food handlers in the Omdurman area of Sudan. J Micbiol Immunol Infect, 2010; 43(1): 70-73.
- Jodi Woan-Fei Law., Nurul-Syakima Ab Mutalib., Kok-Gan Chan., and Learn-Han Lee. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol, 2014; 5: 770.
- Zhao X., Lin C. W., Wang J., and Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechn, 2014; 24: 297–312.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.