ISSN: 0973-7510
E-ISSN: 2581-690X
Aspergillosis is a fungal disease caused by different species of Aspergillus. They live in soil,dust and decomposed material. Number of Aspergillus species found till now is about 300 and more are still to be identified. Only few Aspergillus species can cause human disease and the most common species for human infection is Aspergillus fumigatus, which is a ubiquitous airborne saprophytic fungus. Severity of the disease ranges from an allergic response to life-threatening generalized infection. They grow optimally at 37°C and can grow upto 50°C. The fungal conidia are being constantly inhaled by humans and animals everyday normally gets eliminated by innate immune mechanism. Due to increasing number of immunocompromised patients, severe and fatal Aspergillosis cases have augmented. Currently, available antifungal drug for the treatment of Aspergillosis act on these three molecular target are 14 alpha demethylase for Azoles, ergosterol for Polyene and β-1,3-glucan synthase for Echinocandin. These antifungal drug show high resistance problem and toxicity. So, it is high time to develop new drugs for treatment with reduced toxicity and drug resistant problem. Synthesis of essential amino acid is absent in human as they obtain it from their diet but fungi synthesis these amino acid. Thus, enzymes in this pathway acts as novel drug target. This article summarizes promising drug targets presents in different metabolic pathway of Aspergillus genome and discusses their molecular functions in detail. This review also list down the inhibitors of these novel target. We present a comprehensive review that will pave way for discovery and development of novel antifungals against these drug targets for Aspergillosis treatment.
Aspergillosis, Novel Targets, Aspergillus fumigatus, Antifungal, Pathways, Enzymes, Fungi, Inhibitors
In 1729, Pier Antonio Micheli who was both biologist and priest in Italy, named a mold “Aspergillus” as it looked like a holy water sprinkler (Aspergillum). The genus Aspergillus has approximately 184 species. A total of 40 among them are known to cause human or animal infections. Aspergillus fumigatus is reported as most frequent species for causing human infections.
The physician George W. Fresenius was the first who described the species “fumigatus” in 1863. Aspergillosis occurs worldwide having estimated life-threatening infection per year at that location is >200,000 and thus having the mortality rate of 30-95% in the infected population1. Aspergillus fumigatus is an ubiquitous human fungal pathogen due to its effective dispersal in air, survival and growth in the wide range of environmental condition, swift adaptability to host environment, physical characteristics that allow conidia to reach distal airway2. Aspergillosis includes a wide range of diseases, each related to a spectrum of abnormal immune responses within the host. It is broadly classified into three categories; allergic, chronic and invasive. The pathogen has eight chromosomes comprising 28.526 Mb genome length which codes for 9630 proteins. Due to severity of antifungal agents with the problem of side effect and drug resistance emergence, treatment of fungal infection is confined3,4. For the treatment of Aspergillosis, four different class of antifungal drug is used which includes- azoles, polyenes, echinocandins, and alyllamines5. Since 2006, no new types of antifungal drug have been revealed till now6.
The drug targets with their metabolic pathway and functions
For developing drugs against Aspergillosis many target have been studied and still the hunt is on for discovering drugs with better efficacy and precision. Some of the drug targets have been already explored while other targets are still under investigation for designing inhibitors against them. Such targets have been described in detail along with their biosynthesis pathway,biological role, molecular function and their sub-cellular localisation. A drug target in pathogen must be essential for survival of pathogen and it shall have no significantly similar genes in host genome. The following point we should consider while finding the drug target-(1)Target must be essential for the survival of fungi.(2)Both human and fungi shares basic eukaryotic characteristics,so target or inhibitors should provide favourable therapeutic-toxic ratio.(3)Target of fungal pathogen is widely spread and it should be economically attractive7.
We have summarized 33 novel therapeutic targets present in Aspergillus genome which may further facilitate the antifungal drug discovery.
Amino acid biosynthetic pathway
There are twenty amino acid in which only nine are essential for human,which they cannot synthesize in their body. Thus,some of the steps of the amino acid biosynthetic pathway was catalyzed by the enzymes which is absent in human. So,on targeting these enzymes we find some novel drug targets. In pathogenesis of A.fumigatus,biosynthetic pathways like biosynthesis of lysine, aromatic amino acids and sulfur-containing amino acids such as methionine and cysteine are determinants of its virulence8. Growth defect and reduced virulence factor is seen when there is some defect in amino acid biosynthetic pathway9.
Biosynthesis of Histidine
Histidine biosynthesis pathway is present in bacteria,fungi and plant but it is absent in mammal. Due to this feature, it will be an attractive target for antifungal drug discovery. Histidine biosynthesis pathway is closely linked to the metabolism to purine and pentose rather than any other amino acid. Both bacteria and fungi have similar enzyme and intermediates in histidine biosynthesis pathway but having dissimilar operon and controlling genes10. The novel target in this pathway are as follow-
Imidazole glycerol phosphate dehydratase
In, histidine biosynthesis pathway, imidazole glycerol-phosphate (IGP) synthesizes imidazoleacetol-phosphate (IAP)in the presence of Imidazole glycerol-phosphate dehydratase (IGPD; EC 4.2.1.19)11. In Aspergillus nidulans, erasure of the IGPD encoding gene, named hisB, was known to cause histidine auxotrophy. The IGPD activity containing protein is monofunctional. Removal of the gene encoding imidazole glycerol-phosphate dehydratase (HisB) results in histidine auxotrophy, resistance declines to both starvation and overabundance of different heavy metals, including iron, copper and zinc, which assume an essential part in antimicrobial host defence. Experimentally, it resulted to decrease pathogenicity in pulmonary infection, systemic infection,corneal infection of murine and larvae of wax moth12.
Histidinol dehydrogenase
The terminal step in the biosynthesis of histidine is catalysed in the presence of enzyme histidinol dehydrogenase. HisD gene encode the enzyme Histidinol dehydrogenase13. This enzymes belongs to the family of oxidoreductase14.
L-histidinol + 2NAD↔L-histidine + 2NADH + 2H
In Fungi, histidinol dehydrogenases are multifunctional catalysts, their c-terminal domain which catalyzes three different steps of histidine biosynthesis15. HisD gene is absent in mammals,thus it can be used as target for herbicide development also13.
ATP phosphoribosyl transferase
It catalyse the first step in the biosynthesis of Histidine. This pathway is present only in bacteria, fungi, and plants. ATP-phosphoribosyltransferase (ATP-PRT) is the first enzyme of this pathway. ATP-phosphoribosyltransferase (ATP-PRT) form N’ -5’ -phosphoribosyl-ATP (PR-ATP) on condensation with ATP. There are two form of ATP-PRT,one is long having two catalytic domain and a C-terminal regulatory domain and the other is short where regulatory domin is missing. This pathway is, therefore, an promising potential target for the developing non-toxic antifungals16.
Phosphoribosyl AMP cyclohydrolase
Phosphoribosyl-AMP cyclohydrolase (EC:3.5.4.19) involved in catalyzing the third step of the histidine biosynthetic pathway which requires Zn2+ ions. In which hydrolysis of phosphoribosyl-AMP takes place. It additionallyin volved in incorporating various histidine biosynthesis bifunctional proteins17.
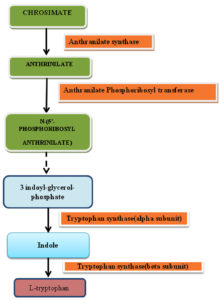
Biosynthesis of Tryptophan
Microorganisms like fungi commonly synthesize tryptophan from shikimic acid or anthranilate19.
Tryptophan synthase
In the biosynthesis of tryptophan, tryptophan synthase an essential enzyme catalyse the last step of the pathway in which indole is converted into L-tryptophan. Tryptophan synthase represents as excellent target for the development of new antifungal agents as genes of tryptophan biosynthesis pathway is absent in human20.
Anthranilate synthase
Anthrinilate is intermediate of tryptophan biosynthesis and it is produce by microorganism which is used by the industry for the synthesis of dyes, perfumes and pharmaceutical compounds.Many prokaryotics microorganisms was engineered to overproduce anthrinilate but on engineering eukaryotic microorganisms production of anthrinilate was less21.
Anthranilate synthase (EC 4.1.3.27) is involved in catalyzing the reaction which prompts the biosynthesis of tryptophan from either chorismate and ammonia or chrosimate and glutamine22. In every single microbial species, anthranilate synthase (AS) is an oligomer of nonidentical subunits assigned AS alpha-subunit (ASI or part I) and AS beta-subunit (ASII or segment II). In a few living beings, the subunits are related and give an αβ dimer and in others an α2 β2 tetramer. In prokaryotes like blue-green algae and bacteria, the trpE gene is encoded by AS subunit while in eukarya like plant and fungi, the TRP2 gene is encoded by ASA1/ASA2 but fl-subunit of the AS enzyme complex contains another enzyme of tryptophan biosynthesis, other genes, besides trpG and trp3 and ASB can encode the multifunctional subunit23. Anthranilate synthase (AS) is responsible for synthesis of anthranilate which is common precursor of many compounds, from chorismate and Gln. Along with chrosimate and Gln, when concentration of ammonia is high then Anthranilate synthase speed up the synthesis step of anthranilate from chorismate in which ammonia acts as an amino donor. Tryptophan cause feedback inhibition of the enzyme24. Thus, on inhibition, this enzyme may put an end to the normal growth and production of pathogen. The reported inhibitors of this enzymes are Acivicin, anthranilic acid, Chanoclavine and Elymoclavine23.
Anthranilate Phosphoribosyl transferase
Anthranilate phosphoribosyltransferase (TrpD) enzyme is member of the family of glycosyltransferases, specifically the pentosyltransferases. It is known for catalyzing the transfer of a phosphoribosyl group to anthranilate which eventually leads to the generation of phosphoribosyl anthranilate. It is involved in tryptophan biosynthesis. This enzyme participates in the phenylalanine, tyrosine and tryptophan biosynthesis and two-component system – general. It has been reported that anthrnilic acid, 3-hydroxyanthranilic acid, n-(5-phospho-d-ribosyl) anthrnilic acid and pyrophosphate are inhibitors of this enzyme. Inhibition of TrpD will be critical for fungal survival as it will stop Tryptophan synthesis23,25,26.
Table (1):
List of novel drug target of Aspergillus fumigatus and their metabolic pathway.
| Novel Drug Target | Metabolic Pathway | ||
|---|---|---|---|
| 1 | Imidazole glycerol phosphate dehydratase | Biosynthesis of Histidine | Amino acid Biosynthetic pathways |
| 2 | Histidinol dehydrogenase | Biosynthesis of Histidine | |
| 3 | ATP phosphoribosyl transferase | Biosynthesis of Histidine | |
| 4 | Phosphoribosyl AMP cyclohydrolase | Biosynthesis of Histidine | |
| 5 | Tryptophan synthase | Biosynthesis of Tryptophan | |
| 6 | Anthranilate synthase | Biosynthesis of Tryptophan | |
| 7 | Anthranilate Phosphoribosyl transferase | Biosynthesis of Tryptophan | |
| 8 | Ketol Acid Reducto-Isomerase(KARI) | Biosynthesis of Valine,leucine and Isoleucine | |
| 9 | Dihydroxy acid dehydratase | Biosynthesis of Valine,leucine and Isoleucine | |
| 10 | Glutamate N-acetyltransferase | Biosynthesis of Arginine | |
| 11 | Homoserine dehydogenase | Biosynthesis of Threonine,Methionine and Lysine | |
| 12 | Orotate phosphoribosyl transferase 1 | Pyrimidine biosynthetic pathway | Other Pathways |
| 13 | Dihydroorotate dehydrogenase(DHODH) | Pyrimidine biosynthetic pathway | |
| 14 | Mannitol-1-phosphate-5-dehydrogenase | Mannitol biosynthesis | |
| 15 | 1,3 beta glucan synthase(G.S.) | Biosynthesis of Starch and sucrose | |
| 16 | N-myristoyltransferase(NMT) | Lipid Biosynthesis | |
| 17 | Phosphatidyl ethanolamine N-methyl transferase | Lipid Biosynthesis | |
| 18 | Ras protein(RasB) | Signal Transduction pathway | |
| 19 | Thiroredoxin reductase | Thioredoxin pathway | |
| 20 | Chrosimate mutase | Shikimate pathway | |
| 21 | 3-deoxy-7-phosphoheptulonate synthase | Shikimate pathway | |
| 22 | Phosphomevalonate kinase | Isoprenoid/Mevalonate pathway | |
| 23 | ACP(Acetyl transferase) | Biosynthesis of fattyacid | |
| 24 | Fatty acid synthase | Biosynthesis of fattyacid | |
| 25 | Hsp90 | Hsp90-calcineurin pathway | |
| 26 | Calcineurin | Hsp90-calcineurin pathway | |
| 27 | Trehalose-6-phosphate synthase | Trehalose Biosynthetic pathways | |
| 28 | Trehalose phosphate phosphatase (Alpha-alpha-trehalose phosphate synthase) | Trehalose Biosynthetic pathways | |
| 29 | 2-methyl isocitrate lyase | Propionate metabolism | |
| 30 | CYP51A | Sterol Biosynthesis Pathway | |
| 31 | Urate oxidase(Uricase) | Purine degradation pathway | |
| 32 | Riboflavin(VITAMIN –B2) | Vitamin Biosynthetic pathway | |
| 33 | 3,4 dihydoxy-2-butanone 4 phosphate synthase | Vitamin Biosynthetic pathway | |
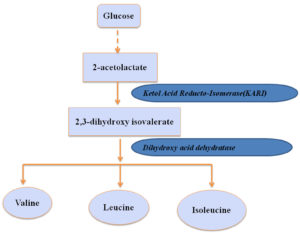
Biosynthesis of Valine, leucine and Isoleucine
In comparison to fungi, humans are unable to synthesize these branched chain amino acids (Valine, leucine and Isoleucine).These biosynthesis pathway has three steps till the production of 2-ketoisovalerate which later breakdown for the synthesis of these branched chain amino acid -valine, leucine, and isoleucine27.
Ketol Acid Reducto-Isomerase(KARI)
Ketol-acid reductoisomerase (KARI) is an enzyme which is involved in the biosynthesis of chain amino acids, Valine, leucine, isoleucine, Pantothenate and CoA in Aspergillus. Metabolization of isoleucine and valine biosynthesis in Aspergillus is done through threonine moiety. For KARI to catalyze a reaction, Mg++ is required as cofactor and NAPDH as a coenzyme. An interruption in this pathway influences the survival of the Aspergillus under the condition of threonine limitation. The interesting fact about this enzyme is its involvement in the biosynthesis of leucine, isoleucine, and valine which is an essential amino acid for a human. In human, this enzyme is not be able to amend the amino acid metabolism on inhibiting but in a pathogen, inhibition occurs when these amino acids are not available23,28. Thus, the KARI is selected as a putative antifungal target. They reported N-hydroxy-n-isopropy loxamate, P-chloromercuribenzoic acid, 2-methyllactic acid and 2-oxo-p-hydroxy isovalerate as inhibitors of KARI23.
Dihydroxy acid dehydratase
The dehydration and tautomerization reaction of two 2,3dihydroxycarboxylic acids (occurs naturally) to the corresponding 2- kaectoids was the third step in the branched chain amino acid biosynthesis which was catalyzed by dihydroxy acid dehydratase. This branched chain amino acid biosynthetic pathway synthesize valine, leucine, CoA, and isoleucine. Thus,the intermediates present in the biosynthesis of valine, leucine, CoA, and isoleucine were used as the drug target. These enzymes were not present within the human host. Therefore, the side effect on the host will be minimum. There are no reported inhibitors23.
Biosynthesis of Arginine
Arginine is converted into NO (nitric oxide) in the presence of enzyme Nitric oxide synthase. Nitric oxide is necessary for fungal growth and development. It also acts as signalling molecules29.
Glutamate N-acetyltransferase
In the biosynthesis of ornithine, transacetylation reaction occurs between N2-acetylornithine and glutamate which is catalyzed by the enzyme Glutamate N-acetyltransferase. It uses acetyl acceptors in the form of arginine, glutamine, and lysine. This protein produce an high intracellular concentration of arginine for biosynthesis of clavam biosynthesis inspite of primary metabolism. It is also involved in the pathway of clavulanate biosynthesis, which is part of Antibiotic biosynthesis30,31.
Biosynthesis of Threonine, Methionine and Lysine
Homoserine dehydogenase
This enzyme belongs to the family of oxidoreductase. The biosynthesis of threonine, isoleucine, and methionine from aspartic acid is catalysed by homoserine dehydrogenase. It is absent in human thus,it can be a antifungal target with high specificity and few side effect32,33.
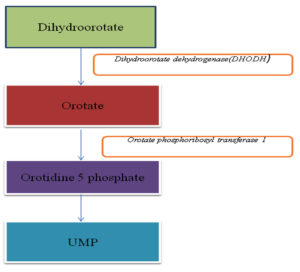
Pyrimidine biosynthetic pathway
Synthesis of uracil monophosphate(UMP) is essential for nucleic acid synthesis. Using pyrimidine, some cell also make UMP. Pyrimidine is also required for cell proliferation and adaptation to cell stress34.
Orotate phosphoribosyl transferase 1
Orotate phosphoribosyltransferase (OPRTase) is the among the top ten phosphoribosyl transferases enzyme, which is required in both de novo and salvage pathway of nucleotide synthesis. This enzyme is also require for histidine and tryptophan formation. Orotate phosphoribosyltransferase (OPRTase) also play essential role in the biosynthesis of pyrimidine nucleotides. De novo pyrimidine nucleotide synthesis pathway leads to the synthesis of the pyrimidine ring earlier to attachment of the ribosyl group. When orotate reacts with ribose-5-phosphate donor 5-phosphoribosyl-1-pyrophosphate (PRPP) it synthesizes orotidine-5-phosphate (OMP) and pyrophosphate in the presence of OPRTase. It signifies essentiality of OPRTase for de novo pyrimidine biosynthesis35,36.
Dihydroorotate dehydrogenase(DHODH)
DHODH is involved in denovo synthesis of pyrimidine and catalyzes the fourth step of pyrimidine biosynthesis pathway in which it converts dihydroorotate to orotate37. On the basis of the difference in amino acid sequence, there are two classes of DHODH. Class II DHODH is found in most of the fungi (including A.fumigatus and C.albicans) animals, plants, gram-negative bacteria and archaebacteria38. They are omnipresent FMN (flavin mononucleotide) flavoenzymes which are involved in both basic and applied area of research. Teriflunomide is known to inhibit DHODH in humans39.DHODH is widely used as a potential drug target in infectious disease, rheumatology, oncology and is a up-and-coming model for the evolution study for enzymatic catalysis40.
Mannitol biosynthesis
Mannitol-1-phosphate-5-dehydrogenase
The NAD-dependent reduction of mannitol-1-phosphates from fructose-6-phosphate is catalyzed by Mannitol dehydrogenase. On dephosphorylation, mannitol-1-phosphates forms Mannitol in the presence of Mannitol-1-p-phosphatase, which is the irreversible step41. Mannitol 2-dehydrogenase then catalyzes the NAD-dependent reduction of mannitol to fructose,which again on phosphorylation form fructose-6-phosphate. Thus,this cycle will run in this direction where Mannitol-1-phosphate-5-dehydrogenase is present. Fungal mannitol serves as carbon source, as reservoir of reducing power, stress resilience and spores dispersal agent. Dehydrogenases are evolutionarily related, including mannitol 2-dehydrogenase (gene mtlK), mannitol-1-phosphate 5-dehydrogenase (gene mtlD) and mannonate oxidoreductase (fructuronate reductase) (gene uxuB)42.
Biosynthesis of Starch and sucrose 1,3 beta glucan synthase(G.S.)
Fungal 1,3 glucan plays an important role in construction of cell wall and growth of cell. Its localization in plasma membrane is essential for its activity. This enzyme form fungal 1,3 b-glucans on polymerization of uridine 5-diphosphoglucose.Fks is a transmembrane protein is the catalytic subunit of GS complex. Fks is located in growing part of cell like cell tip and septum site. On inhibiting this enzyme,there is depletion in 1,3 b-glucans. The reported inhibitors of this enzymes are echinocandin and pneumocandin43.
Lipid Biosynthesis
Lipids such as phospholipids, sphingolipids fatty acids, sterols are important component of cell membrane. They also act as signalling molecules by regulating cellular metabolism44.
N-myristoyltransferase(NMT)
This enzyme catalyzes the process of adding up of myristic acid to the amino-terminal glycine residue in a number of eukaryotic proteins including protozoa, fungi and mammals and viral proteins. Cellular function, signal transduction cascade functions, and membrane targeting function of a protein is NMT dependent45,46. N-myristoylation is a crucial protein to all pathogenic fungi and plays an important role in cell morphology and cell wall integrity. This protein is absent from human cell. Thus NMT is a promising therapeutic target for antiviral, antiparasitics, antifungal and anticancer drugs47-49. Discovery of pyrazole sulphonamide compound as an inhibitor for NMT has been obtained after screening a library and under repressive condition, it has shown fungicidal activity46.
Phosphatidyl ethanolamine N-methyl transferase
In the biosynthesis of phosphatidylcholine, the initial step of the methylation pathway was catalyzed by phosphatidyl ethanolamine N-methyl transferase in which phosphatidylethanolamine (PE) is converted to form phosphatidylcholine (PC). This protein is involved in the pathway of phosphatidylcholine biosynthesis, which is part of Phospholipid metabolism and it is one among two pathways for PC biosynthesis50,51. Methyltransferase show essential role in different cellular processes which includes biosynthesis, signal transduction, repair of protein, regulation of protein and gene silencing52,53.
Signal Transduction pathway
Ras protein(RasB)
Ras protein was located in the plasma membrane as it is a low molecular weight monomeric G-protein. It is required for Ras network interaction which has a crucial role in fungal growth and its virulence. Ras protein induced by external stimuli work as a signal mediator for different downstream cascades and activates transcription factors and play role in different types of a cellular process like cell division, differentiation, growth, and survival.
In a human cell, Ras protein consists of three isoforms (HRas, KRas, NRas) but Aspergillus fumigatus only had two Ras homologs(RasA, RasB). The sequence of RasA is more similar to human HRas with homologs found in most eukaryotes and filamentous fungi only produce RasB. Both protein RasA and RasB alter virulence in A.fumigatus and other pathogenic fungi. Both of them exhibit discrete role but have some common roles in conidial germination, conidiogenesis, mitosis and mycelia growth. In regulating cell response to the various types of stresses from a wide range of effector protein, an important role is played by Ras-mediated signaling pathway. This pathway control host virulence in pathogenic fungi. Thus, this protein and their effector represent a potential target of intervention for novel antifungal therapy54.
Thioredoxin pathway
Thiroredoxin reductase
Thioredoxin (Trx) and thioredoxin reductase (TrxR) are two enzymes of Thioredoxin systems. TrxR is responsible for the reduction of Thireodoxin(Trx). Thioredoxins is tiny protein of 12–13 kDa and is distributed universally. Two categories of Thioredoxin reductase are found in all organisms, one having low molecular weight about 35–36 kDa subunits and the other one is having high molecular weight of 55–58 kDa. Prokaryotes, archaea, plants, and fungi has low molecular weight(TrxR) and high molecular weight (TrxR) is present in higher eukaryotes. High molecular weight TrxR contain an extra redox active site in the C-terminal from low TrxR. This active site is responsible for the interaction with the substrate.They are involved in the regulation of methionine biosynthesis,cell growth,gene transcription and apoptosis55. Number of inhibitors targetting this pathway have been identified which are used as antibacterial and anti-cancer drugs56.
Shikimate pathway
The shikimate pathway is found in both microorganisms and plants but missing in animals57. In the formation of phenylalanine, tyrosine, tryptophan and some aromatic compounds,study of this pathway is essential.
Chrosimate mutase
Chorismate reside in a middle place in the shikimate pathway which connect primary and secondry metabolism in bacteria,plant and fungi58.Conversion of chorismate to prephenate is done by Claisen rearrangement in the presence of chorismate mutase (CM). Enzymes other than chrosimate mutase which utilize chorismate are chorismate lyase, isochorismate synthase, anthranilate synthase, and p-aminobenzoate synthase. CMs exist in various forms both functionally and structurally.This pathway is also absent in human. Thus,chrosimate mutase can be a novel drug target for the development of antifungal drugs51.
3-deoxy-7-phosphoheptulonate synthase
In shikimate pathway, DAHP is be responsible for biosynthesis of phenylalanine, tyrosine, and tryptophan. It regulates the amount of cabon entering the pathway59. When phosphoenolpyruvate (PEP) and D-erythrose-4-phosphate (E4P) condense,it synthesizes 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP). This conversion is catalyzed by DAHP synthase. The enzyme is a target for negative-feedback regulation by pathway intermediates or by their end products60.
Isoprenoid/Mevalonate pathway
Mevalonate pathway is also known as Isoprenoid pathway or HMG-CoA reductase pathway. This pathway consist of seven reactions which starts with acetyl CoA and finally produces isopentyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). Statin inhibit this pathway and thus, no production of mevalonate5-phosphate61,62.
Phosphomevalonate kinase
In the biosynthesis of sterol (fungal ergosterol), mevalonate is the key metabolite and is phosphorylated by mevalonate kinase. This enzyme convert mevalonate 5-phosphate and ATP to mevalonate 5-diphosphate and ADP61.
Biosynthesis of fatty acid
ACP(Acetyl transferase)
The pathway of fatty acid biosynthesis has been well elucidated in both prokaryotes and eukaryotes63. In fungi, synthesis of fatty acid synthesis is divided into different compartment having large multifunctional enzymes: the fatty acid synthases (FASs). There are three different system for the synthesis of fatty acid by de novo are studied. Usually type I fatty acid synthase system (FAS I) was used by eukaryotic and prokaryotic while FAS II was used by bacteria and FAS III was used by some parasite. The fungal fatty acid synthase complex consist of two polyfunctional protein. Due to essential role of fatty acid biosynthesis, this pathway makes the FAS systems potential targets for novel antifungals64,65.
Fatty acid synthase
Fatty acids (FAs) is synthesized in the presence of fatty acid synthase (FAS) from acetyl CoA66. In Aspergillus nidulans, there are two different form of fatty acid synthase. One of them is for the metabolism of primary fatty acid and the other is for secondry metabolism. Primary FAS requires long chain fatty acid for growth and secondry FAS grows normally67. FAS catalyse the synthesis of palmitate from acetylCoA and malonylCoA in the presence of NADPH. Fatty acid are phospholipid part of the cell membraneand play important role in intracellular communication as lipid messenger68.
Hsp90-calcineurin pathway
Calcineurin, lysine deacetylases (KDAC), other client proteins and Hsp90 as a center form a complex network. This complex network, in response to stress occurs due to some antifungal compound coordinates the compensatory repair mechanism of the cell wall.
Hsp90
Hsp90 is also termed as heat shock protein, stress protein or molecular chaperons. They are a group of conserved protein which prevents misfolding and aggregation of the protein. They initiate adaptive response under stress condition69. In Aspergillus fumigatus, Hsp90 consists of 706 amino acids. They are approx 60-65% homologous to human and about 75% with yeast. It is highly conserved and found in all living cells70. Hsp90 control the activity of its linked protein calcineurin in repairing mechanism of the cell wall and in stress response69,71. It is vital in bringing out molecular responses to ecological changes, morphogenesis, resistance occurs due to antifungal compound, and fungal pathogenicity72.
Studies suggests that suppressed Hsp90 causes drastic changes in A. fumigatus conidiation, germination, and hyphal growth . The Hsp90 inhibitor like geldanamycin, when used alone, show restricted antifungal activity. It can not be used for treating fungal disease due to its cross-reactivity against the human proteins and lack of its fungal specificity. The Hsp90 inhibitor geldanamycin when used with FK506 show synergistic effect against echinocandin resistant strain and when it used with caspofungin it shows synergistic effect against the azole-resistant strains, achieving a fungicidal activity73,74.
Calcineurin
Calcineurin is a Calcium and calmodulin- activated protein phosphatase enzyme which is present from fungi to mammals75. Calcineurin(CrzA) is serine/threonine-specific protein phosphatase. In fungi, morphology of cell and its virulence is controlled by CrzA. The activity of transcription factor, CRZ1 and its localization is to be in command of CrzA by dephosphorylating. In A. fumigatus, calcineurin activation is directly proportional to serum growth. When it inactivates,it decreases the virulence and provides decreased filamentation and thus, there is no growth76. In fungi, a critical signal transduction pathway which is essential for growth, morphology, stress response and pathogenicity is Calcineurin pathway. This pathway build a prospect for the development of new antifungal therapies by targeting it77. The function of Calcineurin is inhibited by cyclosporine A (CsA) and tacrolimus (FK506) on binding with their particular intracellular receptor, the immunophilins cyclophilin A (CyA) and FK506-binding protein (FKBP12)78. Calcineurin is essential for fungal development, stress responses, and virulence79. CrzA of Aspergillus fumigatus has approx 14.9% identity with human protein NFAT, minimizing the cross-reactivity possibility with human target, and thus it can be a good antifungal target80.
Trehalose Biosynthetic pathways
Trehalose biosynthetic pathways arises as novel drug target for antifungal drug. Trehalose acts as energy reservoir and under stress condition it produces ATP. It also protect plasma membrane from degradation7. The main feature of this pathway is its direct link with glycolysis and the presence of two enzyme which is trehalose-6-phosphate synthase (Tps1), and trehalose phosphate phosphatase (Tps2)81. Trehalose pathway is absent in human and thus, on targeting, we may get some novel drug target.
Trehalose-6-phosphate synthase
Trehalose is a disaccharide form of glucose and it is present in bacteria, plant, insect and fungi.
In yeast and fungi,trehalose act as reserve carbohydrate other than glycogen. It also play important role in various cellular process like glycolysis, germination and sporulation. It is absent in human thus, it can be considered as novel antifungal target82,83. In stress condition, trehalose helps protein to uphold their native conformation. Trehalose protects cell from a number of unfavourable conditions such as heat, desiccation, freezing84, hydrostatic pressure85, nutrient starvation, enviormental stress and several abiotic stresses86.
Trehalose phosphate phosphatase (Alpha-alpha-trehalose phosphate synthase)
Trehalose, which is a sugar moiety is used as a source of carbon, is essential for survival of conidia. It protects cells against a variety of environmental stresses including desiccation, dehydration, heat, cold and oxidation87. The enzyme alpha-alpha-trehalose phosphate synthase catalyzes the trehalose formation reaction. Firstly, trehalose 6-phosphate is synthesized from UDP glucose (UDPG) and glucose 6-phosphate (G-6-P). Trehalose-6- Phosphatase then converts it into to free trehalose. This sugar is found in a wide variety of organisms. There are no reported inhibitors of Alpha-alpha-trehalose phosphate synthase23.
Propionate metabolism
Propionate is used as source of energy and carbon in microorganisms. Methylcitrate cycle (MCC) is the main propionate metabolizing route in fungi. The cycle starts with propionate and finishes with the α-oxidation of propionate88.
2-methyl isocitrate lyase
Methylisocitrate lyase is main enzymes of the cycle to catalyzes the reaction in which methylisocitrate breaks down into succinate and pyruvate. It shows that disruption of the methylisocitrate lyase leads to an inhibition of growth and reduced conidiation89.
Sterol Biosynthesis Pathway
Sterol biosynthetic pathway of human is mostly similar to fungi, both of them result in two different molecules. Ergosterol is the major sterol present in fungal cell membranes and cholesterol is the major sterol found in mammals90. Studies suggest that Ergosterol is responsible to conserve mitochondrial DNA in fungi91.
CYP51A
In eukaryotes, cytochrome P450 14a-sterol demethylases (CYP51) is the vital enzymes for sterol biosynthesis. CYP51 belongs to the superfamily of haem-containing enzymes, cytochrome P450. CYP51 is situated inside the superficial membrane of the endoplasmic reticulum and it catalyzes methyl group removal from carbon 14 position. Binding of Azole to CYP51 is non-competitive, which causes reduction in the final fungal sterol (usually ergosterol) and simultaneously cause accumulation of 14-methylated sterols which cause disruption of cell membrane by inhibiting fungal growth . Aspergillus species include two CYP51 isoenzymes, ie CYP51A, and CYP51B which is considered as a drug target92,93. The biosynthesis of ergosterol involves approximately 20 enzymes and includes the synthesis of squalene from mevalonate. At a genomic stage, the presence of multiple genes which encodes key enzymes having different 14-α sterol demethylases (Cyp51A and Cyp51B) and three C-5 sterol desaturases (Erg3) was confirmd by examining this pathway in A. Fumigatus. The fungal CYP51 (lanosterol demethylase) was inhibited by the azole class of antifungal drugs through competitive, reversible binding to the heme cofactor which was located in the enzyme active site94.
Purine degradation pathway
The breaking down of purine to uric acid is conserved among different organisms but further degradationof this reaction is depend on the presence of enzyme in different organism.
Uric acid in the presence of uricase form allantonin, which is used as carbon and nitrogen source to plant and fungi95,96.
Urate oxidase(Uricase)
Urate oxidase plays an important role in the degradation of purine pathway. It catalyses the oxidation of uric acid and produces allantoin. Uricase is not expressed in the genome of human and other higher animals and thus, uric acid was the end product of purine degradation. In fungi, purine degrades in its simplest form i.e. ammonia which causes a serious threat to human97,98. Fungal allantoin provide nitrogenous product to the host plant. Reportedly Amelide, 8-azaxanthine, Cyanurate, dicyandiamide, dithionite, glycerol, guanine,2-hydroxypurine, oxonic acid, sucrose and heavy metal ion act as inhibitor of urate oxidase23.
Vitamin Biosynthetic pathway
According to Meir et al., inhibitors of these pathways inhibit many metabolic processes, which can leads to cell death99.
Riboflavin(VITAMIN –B2)
The essential constituent of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) is Riboflavin and in A.fumigatus 238 flavoproteins is used as such. Deletion of this enzyme result in disturbance of many cellular process include iron homeostatis and vitamin auxotrophy100.
3,4 dihydoxy-2-butanone 4 phosphate synthase
The riboflavin biosynthesis pathway is an essential pathway in most of the fungal pathogens101. Absense of this enzyme in human makes it attractive target for the discovery of antimicrobials against microorganisms that are unable to gain adequate riboflavin from their hosts. It catalyzes a stage in the biosynthesis of riboflavin. The conversion of Ribulose 5-phosphate into 3,4-dihydroxy-2-butanone 4-phosphate and formate involve different chemical reaction i.e. enolization, ketonization, dehydration, skeleton rearrangement, and formate elimination102. 3,4-Dihydroxy-2-butanone 4-phosphate synthase produces eight carbon atom to the xylene ring of the vitamin and thus form the building blocks. There are no reported inhibitors of 3,4 dihydoxy-2-butanone 4 phosphate synthase.
Currently Broad-Spectrum Triazoles, Liposomal Amphotericin B, and Echinocandins are being used for Aspergillosis treatment, while increasing resistance against known antifungals is a major concern103,104. Recent advancements in sequencing technologies have helped in easily sequencing genomes of organisms. Availability of various microbial genomes and their different strains in public domain has facilitated the comparative genomics and mutant discovery. Increasing resistance in fungal pathogens like Aspergillus fumigatus against commonly used antifungal is a major concern. Despite of widespread life-threatening infections due to Aspergillus fumigatus, limited treatments options are available. On the other hand significant increase of drug-resistance is becoming a major challenge to treat fungal infections. There is an urgent need to discover novel of antifungal agents with optimum safety and efficacy105,106. However, broad spectrum antifungal drug development is relatively difficult as the fungal pathogens are more closely related to the host. The fungal pathogens are metabolically similar to their mammalian hosts at cellular level therefore offers few pathogen-specific targets107,108. In this review, we explained novel drug targets for Aspergillosis published within the last years. This review summarizes different drug targets in Aspergillus genomes and implications of their inhibition. This study will facilitate further antifungal drug discovery through exploration of new drug targets in fungal genomes.
ACKNOWLEDGMENTS
SS, NSM and SK are thankful to MNNIT, Allahabad for providing necessary infrastructural facilities. SS is highly thankful to Council of Science and Technology Uttar Pradesh for a project fellowship. AM is highly thankful to Council of Science and Technology Uttar Pradesh for a sponsored project.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
AM conceived this project. SS, NSM and SK reviewed literature. All the authors contributed in writing the manuscript.
FUNDING
This work was funded by a research grant from Council of Science and Technology Uttar Pradesh (Grant No. CST/238 dated 12/05/2015).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4(165):165rv13.
Crossref - Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus-what makes the species a ubiquitous human fungal pathogen?. PLoS Pathogens. 2013;9(12):e1003743.
Crossref - Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 2017;3(4):57.
Crossref - Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol. 2017;71:753-775.
Crossref - Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat Rev Drug Discov. 2010;9(9):719-727.
Crossref - Denning DW, Bromley MJ. Infectious disease. How to bolster the antifungal pipeline. Science. 2015;347(6229):1414-1416.
Crossref - Parente-Rocha JA, Bailao AM, Amaral AC, et al. Antifungal resistance, metabolic routes as drug targets, and new antifungal agents: an overview about endemic dimorphic fungi. Mediators Inflamm. 2017:2017:9870679.
Crossref - Alcazar-Fuoli L. Amino acid biosynthetic pathways as antifungal targets for fungal infections. Virulence. 2016;7(4):376-378.
Crossref - Ramakrishnan J, Rathore SS, Raman T. Review on fungal enzyme inhibitors-potential drug targets to manage human fungal infections. RSC Advances. 2016;6(48):42387-42401.
Crossref - Jastrzebowska K, Gabriel I. Inhibitors of amino acids biosynthesis as antifungal agents. Amino Acids. 2015;47(2):227-249.
Crossref - Glynn SE, Baker PJ, Sedelnikova SE, et al. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure. 2005;13(12):1809-1817.
Crossref - Dietl AM, Amich J, Leal S, et al. Histidine biosynthesis plays a crucial role in metal homeostasis and virulence of Aspergillus fumigatus. Virulence. 2016;7(4):465-476.
Crossref - Barbosa JA, Sivaraman J, Li Y, et al. Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase. Proc Natl Acad Sci. 2002;99(4):1859-1864.
Crossref - Grubmeyer CT, Gray WR. A Cysteine residue (cysteine-116) in the histidinol binding site of histidinol dehydrogenase. Biochemistry. 1986;25(17):4778-4784.
Crossref - Nagai A, Ward E, Beck J, et al. Structural and functional conservation of histidinol dehydrogenase between plants and microbes. Proc Natl Acad Sc. 1991;88(10):4133-4137.
Crossref - Lohkamp B, McDermott G, Campbell SA, Coggins JR, Lapthorn AJ. The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition. J Mol Biol. 2004;336(1):131-144.
Crossref - D’Ordine RL, Klem TJ, Davisson VJ. N1-(5 ‘-Phosphoribosyl) adenosine− 5 ‘-Monophosphate Cyclohydrolase: Purification and Characterization of a Unique Metalloenzyme. Biochemistry. 1999;38(5):1537-1546.
Crossref - Sivaraman J, Myers RS, Boju L, et al. Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI. Biochemistry. 2005;44(30):10071-10080.
Crossref - Radwanski ER, Last RL. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. The Plant Cell. 1995;7(7):921.
Crossref - Simeth NA, Kinateder T, Rajendran C, et al. Towards Photochromic Azobenzene-Based Inhibitors for Tryptophan Synthase. Chemistry (Weinheim an der Bergstrasse, Germany). 2021;27(7):2439-2451.
Crossref - Kuivanen J, Kannisto M, Mojzita D, Rischer H, Toivari M, Jantti J. Engineering of Saccharomyces cerevisiae for anthranilate and methyl anthranilate production. Microbial Cell Factories. 2021;20(1):1-12.
Crossref - Tamir H, Srinivasan PR. Studies of the mechanism of anthranilate synthase reaction. Proc Natl Acad Sci. 1970;66(2):547-551.
Crossref - Morya VK, Kumari S, Kim EK. Imperative pathway analysis to identify the potential drug target for Aspergillus infection. International Journal of Latest Trends in Computing. 2011;2(1):178-182.
- Matsui K, Miwa K, Sano K. Two single base-pair substitutions causing desensitization to tryptophan feedback inhibition of anthranilate synthase and enhanced expression of tryptophan gene of Brevibacterium lactofermentum. J Bacteriol. 1987;169(11):5330-5332.
Crossref - Kim C, Xuong NH, Edwards S, Yee MC, Spraggon G, Mills SE. The crystal structure of anthranilate phosphoribosyltransferase from the enterobacterium Pectobacterium carotovorum. FEBS Letters. 2002;523(1-3):239-246.
Crossref - Perveen S, Rashid N, Tang XF, Imanaka T, Papageorgiou AC. Anthranilate phosphoribosyltransferase from the hyperthermophillic archaeon Thermococcus kodakarensis shows maximum activity with zinc and forms a unique dimeric structure. FEBS Open BioVolume. 2017;7(8):1217-1230.
Crossref - Orasch T, Dietl AM, Shadkchan Y, et al. The leucine biosynthetic pathway is crucial for adaptation to iron starvation and virulence in Aspergillus fumigatus. Virulence. 2019;10(1):925-934.
Crossref - Morya VK, Kumari S, Kim EK. Virtual screening and evaluation of Ketol-Acid Reducto-Isomerase (KARI) as a putative drug target for Aspergillosis. Clin Proteomics. 2012;9:1.
Crossref - Zhao Y, Lim J, Xu J, Yu JH, Zheng W. Nitric oxide as a developmental and metabolic signal in filamentous fungi. Mol Microbiol. 2020;113(5):872-882.
Crossref - Kershaw NJ, McNaughton HJ, Hewitson KS, et al. ORF6 from the clavulanic acid gene cluster of Streptomyces clavuligerus has ornithine acetyltransferase activity. Eur J Biochem. 2002;269(8):2052-2059.
Crossref - Tan Q, Zhao X, He H, Zhang J, Yi T. Carbamoyl phosphate synthetase subunit Cpa1 interacting with Dut1, controls development, arginine biosynthesis, and pathogenicity of Colletotrichum gloeosporioides. Fungal Biol. 2021;125(3):184-190.
Crossref - Jacques SL, Nieman C, Bareich D, Broadhead G, Kinach R, Honek JF, Wright GD. Characterization of yeast homoserine dehydrogenase, an antifungal target: the invariant histidine 309 is important for enzyme integrity. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 2001;1544(1):28-41.
Crossref - Bagatin MC, Pimentel AL, Biavatti DC, et al. Targeting the homoserine dehydrogenase of Paracoccidioides species for treatment of systemic fungal infections. Antimicrob Agents and Chemother. 2017;61(9):e00165-17.
Crossref - de Gontijo FA, Pascon RC, Fernandes L, Machado Jr J, Alspaugh JA, Vallim MA. The role of the de novo pyrimidine biosynthetic pathway in Cryptococcus neoformans high temperature growth and virulence. Fungal Genetics and Biology. 2014;70:12-23.
Crossref - Schramm VL, Grubmeyer C. Phosphoribosyltransferase mechanisms and roles in nucleic acid metabolism. Prog Nucleic Acid Res Mol Biol. 2004;78:261-304.
Crossref - Wang X, Ma C, Wang X, Xu P. Orotate phosphoribosyltransferase from Corynebacterium ammoniagenes lacking a conserved lysine. Journal of Bacteriology. 2007;189(24):9030-9036.
Crossref - Oliver JD, Sibley GE, Beckmann N, et al. F901318 represents a novel class of antifungal drug that inhibits dihydroorotate dehydrogenase. Proc Natl Acad Sci. 2016;113(45):12809-12814.
Crossref - Oliver JD, Thain JL, Bromley MJ, Sibley GEM, Birch M. “Dihydroorotate dehydrogenase as antifungal drug target and quinazolinone-based inhibitors thereof.” U.S. Patent No. 2015;9:034:887. Washington, DC: U.S. Patent and Trademark Office.
- Bar-Or A, Pachner A, Menguy-Vacheron F, Kaplan J, Wiendl H. Teriflunomide and its mechanism of action in multiple sclerosis. Drugs. 2014;74(6):659-674.
Crossref - Reis RA, Calil FA, Feliciano PR, Pinheiro MP, Nonato MC. The dihydroorotate dehydrogenases: Past and present. Arch Biochem Biophys. 2017;632:175-191.
Crossref - Honeyman AL, Curtiss R. Isolation, characterization, and nucleotide sequence of the Streptococcus mutans mannitol-phosphate dehydrogenase gene and the mannitol-specific factor III gene of the phosphoenolpyruvate phosphotransferase system. Infect immun. 1992;60(8):3369-3375.
Crossref - Schneider KH, Giffhorn F, Kaplan S. Cloning, nucleotide sequence and characterization of the mannitol dehydrogenase gene from Rhodobacter sphaeroides. Journal of General Microbiology. 1993;139(10):2475-2484.
Crossref - Liu J, Balasubramanian MK. 1, 3-beta-Glucan synthase: a useful target for antifungal drugs. Curr Drug Targets-Infect Disord. 2001;1(2):159-169.
Crossref - Pan J, Hu C, Yu JH. Lipid biosynthesis as an antifungal target. J Fungi. 2018;4(2):50.
Crossref - Towler DA, Adams SP, Eubanks SR, et al. Purification and characterization of yeast myristoyl CoA: protein N-myristoyltransferase. Proc Natl Acad Sci. 1987;84(9):2708-2712.
Crossref - Fang W, Robinson DA, Raimi OG, et al. N-Myristoyltransferase is a cell wall target in Aspergillus fumigatus. ACS Chemical Biology. 2015;10(6):1425-1434.
Crossref - Selvakumar P, Lakshmikuttyamma A, Shrivastav A, Das SB, Dimmock JR, Sharma RK. Potential role of N-myristoyltransferase in cancer. Prog Lipid Res. 2007;46(1):1-36.
Crossref - Hill BT, Skowronski J. Human N-myristoyltransferases form stable complexes with lentiviral nef and other viral and cellular substrate proteins. J Virol. 2005;79(2):1133-1141.
Crossref - Frearson JA, Brand S, McElroy SP, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464(7289):728-732.
Crossref - Pel HJ, de Winde JH, Archer DB, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology. 2007;25(2):221-231.
Crossref - Lim S, Springstead JR, Yu M, Bartkowski W, Schroder I, Monbouquette HG. Characterization of a key trifunctional enzyme for aromatic amino acid biosynthesis in Archaeoglobus fulgidus. Extremophiles. 2009;13(1):191-198.
Crossref - Schnappauf G, Krappmann S, Braus GH. Tyrosine and tryptophan act through the same binding site at the dimer interface of yeast chorismate mutase. J Biol Chem. 1998;273(27):17012-17017.
Crossref - Cho MH, Corea OR, Yang H, et al. Phenylalanine biosynthesis in Arabidopsis thaliana identification and characterization of Arogenate dehydratases. Journal of Biological Chemistry. 2007;282(42):30827-30835.
Crossref - Abdallah QA, Fortwendel JR. Exploration of Aspergillus fumigatus Ras pathways for novel antifungal drug targets. Front Microbiol. 2015;6:128.
Crossref - Thon M, Al-Abdallah Q, Hortschansky P, Brakhage AA. The thioredoxin system of the filamentous fungus Aspergillus nidulans impact on development and oxidative stress response. J Biol Chem. 2007;282(37):27259-27269.
Crossref - Saccoccia F, Angelucci F, Boumis G, et al. Thioredoxin reductase and its inhibitors. Current Protein and Peptide Science. 2014;15(6):621-646.
Crossref - Herrmann KM, Weaver LM. The shikimate pathway. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50(1):473-503.
Crossref - Hubrich F, Muller M, Andexer JN. Chorismate-and isochorismate converting enzymes: versatile catalysts acting on an important metabolic node. Chem Commun. 2021;57:2441-2463.
Crossref - Walker GE, Dunbar B, Nimmo HG, Coggins JR. Evidence for a novel class of microbial 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase in Streptomyces coelicolor A3 (2), Streptomyces rimosus and Neurospora crassa. Microbiology. 1996;142(8):1973-1982.
Crossref - Shumilin IA, Kretsinger RH, Bauerle RH. Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Escherichia coli. Structure. 1999;7(7):865-875.
Crossref - Houten SM, Waterham HR. Nonorthologous gene displacement of phosphomevalonate kinase. Mol Genet Metab. 2001;72(3):273-276.
Crossref - Chatzivasileiou AO, Ward V, Edgar SM, Stephanopoulos G. Two-step pathway for isoprenoid synthesis. Proc Natl Acad Sci. 2019;116(2):506-511.
Crossref - Lowe PN, Rhodes S. Purification and characterization of [acyl-carrier-protein] acetyltransferase from Escherichia coli. Biochem J. 1988;250(3):789.
Crossref - Johansson P, Wiltschi B, Kumari P, et al. Inhibition of the fungal fatty acid synthase type I multienzyme complex. Proc Natl Acad Sci. 2008;105(35):12803-12808.
Crossref - Jenni S, Leibundgut M, Maier T, Ban N. Architecture of a fungal fatty acid synthase at 5 Å resolution. Science. 2006;311(5765):1263-1267.
Crossref - Hutchinson CR, Fujii I. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu Rev Microbiol. 1995;49(1):201-238
Crossref - Brown DW, Adams TH, Keller NP. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc Natl Acad Sci USA. 1996;93(25):14873-14877.
Crossref - Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68(3):501-517.
Crossref - Lamoth F, Juvvadi PR, Steinbach WJ. Heat shock protein 90 (Hsp90): a novel antifungal target against Aspergillus fumigatus. Crit Rev Microbiol. 2016;42(2):310-321.
- Li Z, Srivastava P. Heat‐shock proteins. Current Protocols in Immunology. 2003; 58: A.1T.1-A.1T.6.
Crossref - Juvvadi PR, Lamoth F, Steinbach WJ. Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol Rev. 2014;28(2-3):56-69.
Crossref - Jacob TR, Peres NTA, Martins MP, et al. Heat shock protein 90 (Hsp90) as a molecular target for the development of novel drugs against the dermatophyte Trichophyton rubrum. Front Microbiol. 2015;6:1241.
Crossref - Lamoth F, Juvvadi PR, Gehrke C, Steinbach WJ. In vitro activity of calcineurin and heat shock protein 90 inhibitors against Aspergillus fumigatus azole-and echinocandin-resistant strains. Antimicrob Agents Chemother. 2013;57(2):1035-1039.
Crossref - Lamoth F, Juvvadi PR, Soderblom EJ, Moseley MA, Asfaw YG, Steinbach WJ. Identification of a key lysine residue in heat shock protein 90 required for azole and echinocandin resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 2014;58(4):1889-1896.
Crossref - Fernandes CM, Dasilva D, Haranahalli K, et al. The future of antifungal drug therapy: novel compounds and targets. Antimicrob Agents Chemother. 2021;65(2):e01719-e01720.
Crossref - Soriani FM, Malavazi I, Savoldi M, et al. Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiology. 2010;10(1):12.
Crossref - Cramer RA, Perfect BZ, Pinchai N, et al. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryotic Cell. 2008;7(7):1085-1097.
Crossref - Van Rossum HH, de Fijter JW, van Pelt J. Pharmacodynamic monitoring of calcineurin inhibition therapy: principles, performance, and perspectives. Ther Drug Monit. 2010;32(1):3-10.
Crossref - LeBlanc EV, Polvi EJ, Veri AO, Prive GG, Cowen LE. Structure-guided approaches to targeting stress responses in human fungal pathogens. J Biol Chem. 2020;295(42):14458-14472.
Crossref - Northrop JP, Ho SN, Chen L, et al. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369(6480):497-502.
Crossref - Perfect JR, Tenor JL, Miao Y, Brennan RG. Trehalose pathway as an antifungal target. Virulence. 2017;8(2):143-149.
Crossref - Al-Bader N, Vanier G, Liu H, et al. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect immun. 2010;78(7):3007-3018.
Crossref - Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie van Leeuwenhoek. 1990;58(3):209-217.
Crossref - Van Laere A. Trehalose, reserve and/or stress metabolite?. FEMS Microbiology Letters. 1989;5(3):201-209.
Crossref - Iwahashi H, Nwaka S, Obuchi K. Evidence for contribution of neutral trehalase in barotolerance of Saccharomyces cerevisiae. Appl Environ Microbiol. 2000;66(12):5182-5185.
Crossref - Zahringer H, Thevelein JM, Nwaka S. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol Microbiol. 2000;35(2):397-406.
Crossref - Chen Q, Haddad GG. Role of trehalose phosphate synthase and trehalose during hypoxia: from flies to mammals. J Exp Biol. 2004;207(18):3125-3129.
Crossref - Santos LPA, Assuncao LDP, Lima PDS, et al. Propionate metabolism in a human pathogenic fungus: Proteomic and biochemical analyses. IMA Fungus. 2020;11:1-16.
Crossref - Brock M. Generation and phenotypic characterization of Aspergillus nidulans methylisocitrate lyase deletion mutants: methylisocitrate inhibits growth and conidiation. Appl Environ Microbiol. 2005;71(9):5465-5475.
Crossref - Joffrion TM, Cushion MT. Sterol biosynthesis and sterol uptake in the fungal pathogen Pneumocystis carinii. FEMS Microbiol Lett. 2010;311(1):1-9.
Crossref - Jorda T, Puig S. Regulation of ergosterol biosynthesis in saccharomyces cerevisiae. Genes. 2020;11(7):795.
Crossref - Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci. 2001;98(6):3068-3073.
Crossref - Parker JE, Warrilow AG, Price CL, Mullins JG, Kelly DE, Kelly SL. Resistance to antifungals that target CYP51. J Chem Biol. 2014;7(4):143-161.
Crossref - Alcazar-Fuoli L, Mellado E. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol. 2012;3:439.
Crossref - Hafez RM, Abdel-Rahman TM, Naguib RM. Uric acid in plants and microorganisms: Biological applications and genetics-A review. J Adv Res. 2017;8(5):475-486.
Crossref - Divon HH, Fluhr R. Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiology Letters. 2007;266(1):65-74.
Crossref - Legoux R, Delpech B, Dumont X, et al. Cloning and expression in Escherichia coli of the gene encoding Aspergillus flavus urate oxidase. J Biol Chem. 1992;267(12):8565-8570.
Crossref - Chitty J, Fraser J. Purine acquisition and synthesis by human fungal pathogens. Microorganisms. 2017;5(2):33.
Crossref - Meir Z, Osherov N. Vitamin biosynthesis as an antifungal target. J Fungi. 2018;4(2):72.
Crossref - Dietl AM, Meir Z, Shadkchan Y, Osherov N, Haas H. Riboflavin and pantothenic acid biosynthesis are crucial for iron homeostasis and virulence in the pathogenic mold Aspergillus fumigatus. Virulence. 2018;9(1):1036-1049.
Crossref - Islam Z, Kumar A, Singh S, Salmon L, Karthikeyan S. Structural basis for competitive inhibition of 3, 4-dihydroxy-2-butanone-4-phosphate synthase from Vibrio cholerae. J Biol Chem. 2015;290(18):11293-11308.
Crossref - Liao DI, Calabrese JC, Wawrzak Z, Viitanen PV, Jordan DB. Crystal structure of 3, 4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure. 2001;9(1):11-18.
Crossref - Jenks JD, Hoenigl M. Treatment of Aspergillosis. J Fungi (Basel, Switzerland). 2018;4(3):98.
Crossref - Roemer T, Krysan DJ. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med. 2014;4(5):a019703-a019703.
Crossref - Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336(6082):647-647.
Crossref - Perfect JR. The antifungal pipeline: a reality check. Nat Rev Drug Discov. 2017;16(9):603-616.
Crossref - Kohler JR, Casadevall A, Perfect J. The spectrum of fungi that infects humans. Cold Spring Harb Perspect Med. 2014;5(1):a019273.
Crossref - Butts A, Krysan DJ. Antifungal drug discovery: something old and something new. PLoS Pathog. 2012;8(9):e1002870.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.