ISSN: 0973-7510
E-ISSN: 2581-690X
Cholera in Odisha’s tribal and coastal areas has been a persistent problem due to Vibrio cholerae O1/O139 for the past 30 years. This study investigates diarrheal outbreaks reported between July to November 2022 from five tribal regions of Odisha; aiming to identify the causative pathogen, its antibiogram profile and virulence genes. Standard techniques were used to culture rectal swabs taken from patients with diarrhea and samples of water to isolate the pathogen, its antibiogram profiles and PCR assays (simplex/multiplex) were used to detect different toxic genes. The heavy rainfall reported in July 2022 led to the spread of the diarrheal disease from Rayagada to neighbouring districts. The V. cholerae O1 Ogawa El Tor, characterised by the ctxB7 genotype and exhibiting multidrug resistance, were identified. Demographic analysis revealed that people aged 14-40 and over 40 years were the most affected with equal distribution among males and females. A high proportion of strains (ranging from 80% to 100%) tested positive for various virulence and drug-resistance genes. The findings offer important insights into the ctxB7 genotypes of V. cholerae O1 strains responsible for outbreaks in Odisha, which will aid in the development of improved prevention and control strategies for future cholera outbreaks. The study also suggests that there might be a possible linkage between the strains in Odisha and those from the recent cholera outbreak in Bangladesh, which can be known through whole genome sequencing after collaboration in future.
Cholera Outbreak, Vibrio cholerae O1, ctxB7, Antibiotic Resistance, Tribal Areas, Odisha
Cholera is a serious gastrointestinal illness resulting from the toxigenic bacterium Vibrio cholerae, primarily spread through fecal-oral transmission via contaminated food and water, as well as poor sanitation practices. The infection can rapidly cause severe dehydration, metabolic acidosis, and death if untreated. Every year, between 1.3 and 4.0 million cases of cholera occur worldwide, leading to between 21,000 and 143,000 fatalities.1 Although there are more than 200 serogroups of V. cholerae based on somatic “O” antigens, only O1 and O139 have been connected to significant outbreaks in emerging and impoverished nations. The O1 serogroup is separated into three biotypes: classical, El Tor, and El Tor variants. Genetic changes in the ctxB gene, which codes for the cholera toxin B component, result in ctxB1 to ctxB12 variants, which are referred to as El Tor variants.2 Cholera outbreaks worldwide have recently been driven by the Haitian form (ctxB7) of V. cholerae O1.3 The first outbreak due to the ctxB7 genotype was reported in Haiti in 2010 after its emergence from Odisha following the 1999 super cyclone.4 Subsequently, this ctxB7 variant has been associated with outbreaks in several countries, including Afghanistan, Bangladesh, Nepal, Yemen, Nigeria, Ethiopia, and others. The 2022 cholera outbreak in Bangladesh was the second-largest since 2000, with 42,000 cases reported between March and April. Similarly, Pakistan reported over 2000 diarrhea cases and 290 confirmed cholera cases in April-May 2022.5 In Odisha, cholera outbreaks occur nearly every three years. Over the last decade, Vibrio cholerae O1 with the ctxB7 genotype and resistance to multiple drugs has caused outbreaks in 2007, 2010, 2012, 2014, 2016, and 2019, particularly in the tribal areas of Odisha.6 This study reports and investigates the diarrheal outbreaks in five tribal districts (Rayagada, Koraput, Nuapada, Subarnapur, and Nabarangpur) of Odisha from July to November 2022, focusing on the causative pathogen, its antibiogram profile, and virulence genes.
Study area
A group from the ICMR-Regional Medical Research Centre’s Microbiology Department in Bhubaneswar, conducted a comprehensive study on the diarrheal outbreak reported in five tribal districts of Odisha, i.e. Rayagada, Koraput, Nabarangpur, Subarnapur, and Nuapada between July and November 2022. The team gathered crucial information on the outbreak, including index cases and details of every diarrhea patient, including their symptoms and medical histories. The spread of infections across different blocks was analysed, with index cases traced through discussions with villagers and hospital records. The study also examined the outbreak’s cause, sources of potable water, chlorination details, and the hygienic conditions in affected villages.
Sample collection
Swabs from diarrhea patients were collected at Community Health Centres (CHCs) and Primary Health Centres (PHCs) in affected villages across five districts (Rayagada, Nuapada, Nabarangpur, Kalahandi, and Koraput) after obtaining informed consent and before administering antibiotics (Figure 1). The swabs were transported in Cary-Blair medium to laboratory for further analysis. On the basis of age, patients were classified into age groups: 0-5 years, over 5 to 14 years, over 15 to 40 years, and above 40 years. Additionally, samples of environmental water were gathered from streams, rivers, tube wells, and household stored water sources used for bathing, cooking, drinking, and cleaning.
Sample processing
Swabs collected from patients and water samples collected from different environmental sources were enriched in APW (alkaline peptone water) and then streaked on TCBS agar, MacConkey agar, and Hektoen enteric agar (BD, USA). Plates were incubated at 37 °C for overnight. Moist yellow colonies were further confirmed serologically by slide agglutination using specific antisera for V. cholerae serogroups (polyvalent O1) and serotypes (monovalent Ogawa/Inaba; BD, USA). Genomic DNA was extracted by the snap chill method for molecular analysis. Pathogen isolation and identification followed established laboratory practices consistent with WHO guidelines.4,7
Antibiotic susceptibility assay
The antibiotic sensitivity test of V. cholerae O1 was assessed on MHA (Muller-Hinton Agar) using the disc diffusion method, following CLSI guidelines and our lab practices.8,9 Fifteen antibiotics were tested: chloramphenicol (C), ciprofloxacin (CIP), azithromycin (AZM), gentamicin (GEN), ampicillin (AMP), tetracycline (TE), streptomycin (S), nalidixic acid (Na), erythromycin (E), ofloxacin (OF), doxycycline (DOX), norfloxacin (NX), cotrimoxazole (COT), furazolidone (FR), and polymyxin B (PB) (BD, USA). Susceptibility was interpreted using standard guidelines and breakpoints.
PCR (Polymerase Chain Reaction) assays
Phenotypically identified V. cholerae strains underwent simplex and multiplex PCR for confirmation and virulence gene detection. Two multiplex PCRs (mPCRs) were performed: mPCRI confirmed V. cholerae using primers for toxin coregulated pilus (tcpA), O1 somatic antigen (rfbO1), zonula occludens toxin (zot), and outer membrane protein (ompW). mPCRII detected additional virulence genes for accessory cholera enterotoxin (ace), hemolysin (hlyA), repeats-in-toxin protein (rtx), toxin regulator (toxR), and outer membrane protein (ompU).10 The strains were also evaluated for antibiotic resistance genes encoding trimethoprim (dfrA1), sulfamethoxazole (sulII), the SXT element and streptomycin (strB).11 Following separation on a 1.8% agarose gel, PCR products were stained with ethidium bromide and examined using a gel documentation system (Bio-Rad, USA).
Mismatch amplification mutation assay (MAMA), Double mismatch amplification mutation assay (DMAMA), PCR & detection of tcpA genes
Using particular primers, MAMA and DMAMA PCR tests were utilised to distinguish between the Haitian (ctxB7), El Tor (ctxB3), and classical (ctxB1) genotypes in all V. cholerae isolates.12,13 Targeting the Classical, El Tor, and Haitian tcpA gene variants, PCR was used to validate biotype-specific characterisation. Two distinct forward primers for each tcpA variant and a common reverse primer for Haitian and El Tor tcpA were employed.14
In the Rayagada district, Kashipur CHC and Tikiri PHC and reported an unexpected spike in diarrhoea cases in July 2022. The first severe case, reported on 7th July in Dudukabahal village, involved a patient with cholera symptoms who was treated and discharged but succumbed on 9th July after symptoms reappeared. Following persistent heavy rainfall in early July, which likely led to water contamination in downstream, more cases emerged in nearby villages like Tikiri and Kakudipadar. The outbreak eventually spread to other blocks, including Kalyansinghpur, Kolnara, and Rayagada Urban. Between July 7, 2022, and November 3, 2022, 414 cases and 10 fatalities were documented; no more cases were reported after that date.
A diarrheal outbreak was documented in Dasmantapur (74 cases from 12th to 27th July 2022) and Laxmipur block of Koraput district (complete line listing data was not available). Field investigations indicated that individuals from Rayagada district contributed to the spread of the outbreak after migrating to these areas.
Additionally, a 70-year-old woman from Nandahandi village, Nabarangpur district, developed diarrhea after attending to a patient in Dasmantampur. A total of 88 cases and one death were reported in Nabarangpur district between July and August 2022, with 44 cases in Khadiaguda, Pitakumili, and other nearby villages. V. cholerae O1 was isolated from the river water in Tulasipadar village, suggesting it as a possible infection source.
In Jatgad village, Nuapada district, another outbreak occurred. The index case, a 70-year-old woman, passed away on July 26, 2022. Diarrhea cases were reported in Majhipada and Harijanpada villages, totaling 84 cases and 1 death between July 25 and September 22, 2022. Tubewell water in Jatgad tested positive for V. cholerae O1 Ogawa.
In Subarnapur district, there was another diarrhea outbreak in Digsira village (index case reported on July 23, 2022) and Suryamunda village (index case reported on July 25, 2022). Contaminated water sources were likely the cause of outbreak. A total of 64 diarrhea cases occurred between July 23 and July 31, 2022 (Figure 2).
Figure 2. Incidence of severe diarrhea cases in 5 tribal districts of Odisha, during July 07, 2022- November 04, 2022
Bacteriological analysis of rectal swabs and water samples
V. cholerae O1 Ogawa was found to be the main pathogen in 77 water samples and 414 rectal swabs that were subjected to bacterial investigation. Antisera testing verified that 114 (27.53%) of the rectal swabs were positive. Positive water samples included three from Rayagada (river, household, hilltop), one from Nabarangpur (river), and one from Nuapada (tube well) (Table).
Table:
Bacteriological analysis of rectal swabs and water samples
| Rectal | Total Rectal Swabs tested | 414 |
| Swabs | Culture Positive samples | 341 (82.4%) |
| E. coli | 225 (54.34%) | |
| V. cholerae O1 Ogawa | 114 (27.53%) | |
| Salmonella spp. | 0 (%) | |
| Shigella spp. | 2 (0.48%) | |
| Culture Negative Samples | 73 (17.63%) | |
| Water | Total Water Samples tested | 77 |
| Samples | No. positive for V. cholerae O1 | 5 (6.5%) [River, supply water from the hill top, tube well and household water] |
Antimicrobial susceptibility patterns
The antibiotic resistance profiles of all V. cholerae O1 strains were tested against 15 different antibiotics using the disc diffusion method, and interpretation was based on the observed zone of inhibition sizes. All V. cholerae isolates showed resistance to ampicillin (100%), furazolidone (100%), and nalidixic acid (100%). Additionally, high levels of resistance were observed for streptomycin (98%), cotrimoxazole (94%), erythromycin (91%), chloramphenicol (79%), and polymyxin B (69%). The V. cholerae O1 isolates from the five cholera affected districts exhibited common sensitivity to ciprofloxacin (70%), norfloxacin (90%), gentamicin (85%), tetracycline (90%), ofloxacin (90%), doxycycline (85%) and azithromycin (82%).
Demographic analysis
Three cholera-affected areas (apart from Koraput) had their diarrhoeal patients demographically analysed. Four age categories of patients were identified: 0-5, 5-14, 14-40, and above 40. Most impacted were those aged 14 to 40, then those over age group 40. The infection rates were lower in the youngest groups 0-5 and 5-14. In every age category, the ratio of male to female patients was about equal (1:1.03).
Prevalence of virulence associated genes
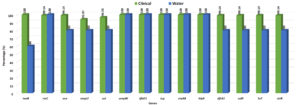
Multiplex PCR experiments were used to identify the genes linked to virulence in isolates of V. cholerae O1. Every isolate of V. cholerae tested positive for the ompW and rfbO1 genes, confirming the species and O1 serogroup. From these five areas, nearly all of the clinical V. cholerae isolates tested positive for every virulence and regulatory gene. All of the V. cholerae isolates had the repeat in toxin rtxC (99.17%), haemolysin hlyA (100%) and cholera toxin ctxAB (100%) present. However, the V. cholerae isolates were 98.35% positive for the accessory cholera enterotoxin (ace), 95.87% positive for the zonula occludens toxin (zot), 100% positive for toxin corregulated pilus (tcp), toxR 97.52% and ompU 93.39% positive gene respectively (Figure 3).
Figure 3. Percentage of virulence and drug-resistance genes in clinical and water isolates of V. cholerae O1 strains isolated from different tribal districts
Profiling of antibiotic resistant genes
The majority of the strains tested positive for antibiotic resistance genes using the PCR assay, which amplified a 278 bp fragment of dfrA1, 515 bp of strB, 626 bp of sulll, and 1035 bp of SXT element. All of these genes may have contributed to the resistance of V. cholerae O1 strains to nalidixic acid, ampicillin, furazolidone, streptomycin, neomycin and cotrimoxazole. 95% of V. cholerae O1 strains had positive results for all of the aforementioned genes (Figure 3).
MAMA and DMAMA PCR assays & detection of tcpA genes
The MAMA and DMAMA PCR assays were conducted for all the V. cholerae O1 isolates. Out of 119 strains 60 strains were randomly tested for the detection of ctxB alleles. All were positive for ctxB7 allele of Haitian variant. Additionally, the existence of the Haitian type tcpA allele was confirmed by the results of the PCR assay for the identification of the tcpA gene (167 bp).
Cholera is still a major global public health concern that draws attention to disparities in social progress. Cholera epidemics in underdeveloped nations are frequently associated with a lack of access to clean drinking water and basic sanitation.15 The WHO estimates that implementing these measures could reduce diarrheal cases by 35%. The absence of safe drinking water and proper sanitation facilities are primary causes of cholera in affected villages. The Indian subcontinent accounts for 78% of reported cholera cases, with India experiencing multiple outbreaks as reported by WHO in 2007.16 In Odisha, cholera has been a persistent challenge, particularly in rural areas where water from unprotected wells and bore wells, prone to contamination, contributes to the spread of waterborne diseases. Acute diarrhoea epidemics and outbreaks are common in rural regions of tropical developing nations where water is obtained from bore wells, chua (shallow pits in rice fields or riverbeds), and unprotected open wells. These sources are vulnerable to groundwater pollution following rains. The water source contamination through open wells and tube wells contributes to the transmission of waterborne diseases.17
This epidemic serves as a compelling example of cholera transmission through both contaminated water sources including poor sanitation practices, inadequate hygiene conditions, and the migration of individuals from outbreak areas—such as patients migrated from Rayagada district to previously unaffected regions like Dasmantapur in Koraput and then to Nabarangpur. The initial detection of the outbreak in the Kashipur block, situated at a higher altitude of Rayagada district highlights the vulnerability of such areas. Subsequently, cholera cases were reported in other blocks of Rayagada district which are located at lower altitudes and downstream of the river originating in the Kashipur block. The geographical gradient with higher-altitude areas being affected first spreading to lower altitude suggests a potential link between altitude and the spread of the disease. Furthermore, an analysis of the index cases in the affected districts reveals a common pattern: individuals visiting neighbouring cholera-affected districts. During the cholera outbreak of 2010 in this region, similar reports were also recorded, which affected Kashipur, Kalyansinghpur, Bissam Cuttack of Rayagada and Mohana of Gajapati district, i.e. affecting from higher altitude to lower altitude.18 These finding underscores the role of human movement and spread of the diarrhea in this region also. It is interesting to note that the incidence of severe diarrhea cases in Rayagada revealed that cases were persistent from July, to November 2022 covering five months which was contrast to the cholera outbreaks of 2007, 2010, 2012 in the same district. It indicated that V. cholerae was existing whether in the environment or in the community for a longer period.
In the present study, the analysis of age groups revealed that the most affected group was aged 14-40 years, followed by those above 40 years. However, in the present study, there was no significant difference between males and females, with a ratio of approximately 1:1.03. This is surprising, as our earlier studies from these tribal areas have shown that females tend to be more affected than males. These findings are consistent with a study conducted in Kathmandu, Nepal, which reported a similar pattern of higher susceptibility among the adult age group.19 Similar results were also observed from Pakistan.20 However, contrasting results were reported from PGIMER, Chandigarh, where the age groups of 5-14 and 14-40 years were found to be most infected, and the male-to-female ratio was 120:71.21 Similarly, contradicting studies were reported from Nepal and Nigeria.22,23
Resistance to ampicillin, furazolidone, nalidixic acid, streptomycin, co-trimoxazole, erythromycin, chloramphenicol, and polymyxin B was shown by the V. cholerae O1 strains that were identified during this cholera outbreak. But those showed susceptibility to azithromycin, doxycycline, ciprofloxacin, gentamicin, norfloxacin, tetracycline, and ofloxacin. A recent study conducted in Kerala in 2021 also reported sensitivity of V. cholerae strains to ciprofloxacin, gentamicin, ofloxacin, and norfloxacin.24 In contrast, a study from Chandigarh reported a resistance rate of 12.72% towards ciprofloxacin.25 The reduced sensitivity to ciprofloxacin observed in our study might be attributed due to its indiscriminate use during the COVID-19 pandemic. Similar findings regarding decreased susceptibility to fluoroquinolones (ciprofloxacin and norfloxacin) have been reported since 1996.26-28 Comparable resistance profiles have also been reported from Solapur,29 Maharashtra,30 Nepal28 and Mozambique.31,32 Previous studies conducted in Odisha have also documented reduced sensitivity towards fluoroquinolone antibiotics.33 The current antibiogram profile of resistance is consistent with previous cholera outbreaks reported in different years in Odisha.4 The resistance of V. cholerae O1 strains to various antibiotics, such as co-trimoxazole, streptomycin and trimethoprim indicates the acquisition of SXT into the chromosome which lead to multidrug-resistance.34 The indiscriminate use of antibiotics in various sectors, including agriculture has contributed to the emergence of multidrug-resistant strains, posing significant challenges for antibiotic therapy worldwide. The majority of the V. cholerae O1 isolates had antibiotic resistance genes, which were subsequently verified by the multiplex PCR experiment.
The pathogenesis of cholera in V. cholerae involves the coordinated action of multiple genes. To assess the toxigenicity and pathogenicity of the V. cholerae isolates in this study, two sets of multiplex PCR (mPCR) were performed. The mPCR II showed that all isolates tested positive for the rtxC, ompU, ace and toxR genes, whereas the mPCR I verified the presence of the ompW, ctxAB, rfbO1 and tcpA genes. The bacteria were identified as V. cholerae by the presence of the ompW gene, and their serogroup was verified as O1 by the presence of the rfbO1 gene. The presence of cholera toxin (CT), a critical marker among the many toxins generated by V. cholerae that were found in this investigation, was indicated by the detection of the toxR gene.34 Gram-negative bacteria are frequently distinguished from other types of bacteria using the RTX toxin, a crucial pathogenicity component. The presumptive cytotoxin (rtxA) and an acetyltransferase (rtxC) are encoded by the RTX toxin gene in V. cholerae. It is also linked to the ATP binding cassette transporter system, which is physically connected to the core element of V. cholerae genome.35,36 Other genes, including toxR and ace, were also examined as part of this epidemic study; the results showed that all V. cholerae O1 isolates had the core toxin region. Several studies from Kerala,30 Chandigarh,21 and Kenya have revealed similar results.37
The ctxB gene in V. cholerae O1 exhibits considerable genetic diversity due to point mutations in the nucleotide sequences, resulting in different amino acid variations. There are twelve distinct genotypes of ctxB associated with various serogroups of V. cholerae.2 Among these genotypes, ctxB7 differs from ctxB1 due to a point mutation at nucleotide position 58 (C to A), leading to a change in the 20th amino acid from Histidine to Aspartic acid.13 In the present study, all isolated strains were confirmed to have ctxB7 genotypes which have also been previously isolated from Odisha during the years 1999, 2007 to 2010, 2012, 2014, 2016, and 2019.6 Similar findings have been reported by other researchers in West Bengal,14 Bihar and Southern India.28 Furthermore, V. cholerae O1 strains with ctxB7 genotypes have been detected in other regions of India, including Chennai, Hyderabad, Solapur and Assam.38-40 It is noteworthy that the same Haitian variant of V. cholerae O1, which caused the devastating cholera epidemic in Haiti in 2010, is believed to have first originated in Odisha in 1999 and was later reported in Nepal.2,4 The ctxB7 genotypes strain of V. cholerae O1 has subsequently might have spread to Africa, Yemen (2015-2017) and most recently reported from Bangladesh in 2022.5
The present study established that the cholera epidemic in the five districts of Odisha were caused by V. cholerae O1 Ogawa with ctxB7 genotype; which might be similar to the strains that caused large outbreak in Bangladesh and Pakistan in April, 2022. It is apprehended that this ctxB7 genotype might have linkage with the recent epidemic of cholera outbreak which will know through whole genome sequencing. Furthermore, the study made it clear that the primary source of V. cholerae that caused cholera outbreaks in Odisha’s tribal communities was environmental water bodies. Effective surveillance and response mechanisms, which are frequently subpar in poor nations, are necessary to control cholera epidemics. The necessity of an efficient active monitoring system with capacity building to identify and contain cholera outbreaks at the right time and location is therefore highlighted by this study. To avoid and control future diarrhoeal epidemics in Odisha’s tribal districts, public cleanliness and the provision of safe drinking water should be given top priority.
ACKNOWLEDGMENTS
The authors sincerely thank the Director, Directorate of Public Health, Government of Odisha, as well as the Chief District Medical Officers (CDMOs) of Rayagada, Nabarangpur, Subarnapur, and Nuapada districts for their kind cooperation in facilitating data collection. Their support in the collection of rectal swabs from diarrheal patients in hospitals and affected villages, along with water samples from impacted areas, is gratefully acknowledged.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analysed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Ali M, Nelson AR, Lopez AL, Sack DA. Updated Global Burden of Cholera in Endemic Countries. PLOS Negl Trop Dis. 2015;9(6):e0003832.
Crossref - Ramamurthy T, Mutreja A, Weill FX, Das B, Ghosh A, Nair GB. Revisiting the Global Epidemiology of Cholera in Conjunction With the Genomics of Vibrio cholerae. Front Public Health. 2019;7:203.
Crossref - Weill FX, Domman D, Njamkepo E, et al. Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature. 2019;565(7738):230-233.
Crossref - Pal BB, Behera DR, Nayak SR, Nayak AK. Origin and Dissemination of Altered El Tor Vibrio cholerae O1 Causing Cholera in Odisha, India: Two and Half Decade’s View. Front Microbiol. 2021;12:757986.
Crossref - World Health Organization. Disease outbreak News Cholera – Global situation. 2022. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON426.
- Pal BB, Nayak AK, Nayak SR. Emergence and spread of different ctxB alleles of Vibrio cholerae O1 in Odisha, India. Int J Infect Dis. 2021;105:730-732.
Crossref - World Health Organization. Laboratory Biosafety Manual. 3rd ed. World Health Organization. 2004.
- Nayak AK, Nayak SR, Dipti Ranjan Behera, Pal BB. Dissemination of Vibrio cholerae O1 isolated from Odisha, India. Environ Microbiol Rep. 2021;13(3):355-363.
Crossref - CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 3rd ed. CLSI guideline M45. Wayne, PA: Clinical and Laboratory Standards Institute. 2016.
- Kumar P, Jain M, Goel AK, et al. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J Med Microbiol. 2009;58(2):234-238.
Crossref - Ramachandran D, Bhanumathi R, Singh DV. Multiplex PCR for detection of antibiotic resistance genes and the SXT element: application in the characterization of Vibrio cholerae. J Med Microbiol. 2007;56(3):346-351.
Crossref - Morita M, Ohnishi M, Arakawa E, et al. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol. 2008;52(6):314-317.
Crossref - Naha A, Pazhani GP, Ganguly M, et al. Development and Evaluation of a PCR Assay for Tracking the Emergence and Dissemination of Haitian Variant ctxB in Vibrio cholerae O1 Strains Isolated from Kolkata, India. J Clin Microbiol. 2012;50(5):1733-1736.
Crossref - Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. Genetic Traits of Vibrio cholerae O1 Haitian Isolates That Are Absent in Contemporary Strains from Kolkata, India. PLOS ONE. 2014;9(11):e112973-e112973.
Crossref - Brick T, Primrose B, Chandrasekhar R, Roy S, Muliyil J, Kang G. Water contamination in urban south India: household storage practices and their implications for water safety and enteric infections. Int J Hyg Environ Health. 2004;207(5):473-480.
Crossref - Cholera 2006. Wkly Epidemiol Rec. 2007;82:273-84.
- Sarkar R, Prabhakar AT, Manickam S, et al. Epidemiological investigation of an outbreak of acute diarrhoeal disease using geographic information systems. Trans Roy Soc Trop Med Hyg. 2007;101(6):587-593.
Crossref - Kar SK, Pal BB, Khuntia HK, Achary KG, Khuntia CP. Emergence and Spread of Tetracycline resistant Vibrio cholerae O1 El Tor variant during 2010 cholera epidemic in the tribal areas of Odisha, India. Int J Infect Dis. 2015;33:45-49.
Crossref - Shrestha UT, Adhikari N, Maharjan R, et al. Multidrug resistant Vibrio cholerae O1 from clinical and environmental samples in Kathmandu city. BMC Infect Dis. 2015;15(1):104.
Crossref - Zahid QA, Nazia Khursheed, Adnan F, Zafar A. Cholera Outbreak 2022 in Karachi: A Report on Serotype and Antibiotic Susceptibility Pattern. J Coll Physicians Surg Pak. 2022;32(12):1613-1616.
Crossref - Taneja N, Mishra A, Batra N, Gupta P, Mahindroo J, Mohan B. Inland cholera in freshwater environs of north India. Vaccine. 2020;38(Suppl 1):A63-A72.
Crossref - Rijal N, Acharya J, Adhikari S, et al. Changing epidemiology and antimicrobial resistance in Vibrio cholerae: AMR surveillance findings (2006-2016) from Nepal. BMC Infect Dis. 2019;19(1):801.
Crossref - Luke GO, Luke A, Ogbondah BO, Nwadiuto I, Abikor V, Owhondah E. Outbreak investigation of cholera in a rural community, Rivers State Nigeria: An interventional epidemiological study. Int J Commun Med Public Health. 2023;10(2):860-868.
Crossref - Reethy PS, Lalitha KV. Characterization of V. cholerae O1 biotype El Tor serotype Ogawa possessing the ctxB gene of the classical biotype isolated from well water associated with the cholera outbreak in Kerala, South India. J Water and Health. 2021;19(3):478-487.
Crossref - Gupta P, Modgil V, Kant V, et al. Phenotypic and genotypic characterization of antimicrobial resistance in clinical isolates of Vibrio cholerae over a decade (2002-2016). Indian J Med Microbiol. 2022;40(1):24-29.
Crossref - Garg P, Sinha S, Chakraborty R, et al. Emergence of Fluoroquinolone-Resistant Strains of Vibrio cholerae O1 Biotype El Tor among Hospitalized Patients with Cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45(5):1605-1606.
Crossref - Krishna BVS, Patil AB, Chandrasekhar MR. Fluoroquinolone-resistant Vibrio cholerae isolated during a cholera outbreak in India. Trans R Soc Trop Med Hyg. 2006;100(3):224-226.
Crossref - Goel AK, Jain M, Kumar P, Jiang SC. Molecular characterization of Vibrio cholerae outbreak strains with altered El Tor biotype from southern India. World J Microbiol Biotechnol. 2009;26(2):281-287.
Crossref - Jain M, Goel AK, Bhattacharya P, Ghatole M, Kamboj DV. Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Tropica. 2011;117(2):152-156.
Crossref - Kumar P, Mishra DK, Deshmukh DG, et al. Haitian variant ctxB producing Vibrio cholerae OI with reduced susceptibility to ciprofloxacin is persistent in Yavatmal, Maharashtra, India, after causing a cholera outbreak. Clin Microbiol Infect. 2014;20(5):O292-O293.
Crossref - Mandomando I, Espasa M, Valles X, et al. Antimicrobial resistance of Vibrio cholerae O1 serotype Ogawa isolated in Manhiחa District Hospital, southern Mozambique. J Antimicrob Chemother. 2007;60(3):662-664.
Crossref - Dengo-Baloi LC, Sema-Baltazar CA, Manhique LV, Chitio JE, Inguane DL, Langa JP. Antibiotics resistance in El Tor Vibrio cholerae 01 isolated during cholera outbreaks in Mozambique from 2012 to 2015. PLoS ONE. 2017;12(8):e0181496.
Crossref - Khuntia HK, Samal SK, Sahoo RK, Kar SK, Pal BB. Escalating emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 and O139 among hospitalized patients with cholera in Orissa, India. J Pure Appl Microbiol. 2009;3(2):811-814.
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910-1914.
Crossref - Kaper JB, Morris JG, Levine MM. Cholera. Clin Microbiol Rev. 1995;8(1):48-86.
Crossref - Coote JG. Structural and functional relationships among the RTX toxin determinants of Gram-negative bacteria. FEMS Microbiol Lett. 1992;88(2):137-162.
Crossref - Bundi M, Shah MM, Odoyo E, et al. Characterization of Vibrio cholerae O1 isolates responsible for cholera outbreaks in Kenya between 1975 and 2017. Microbiol Immunol. 2019;63(9):350-358.
Crossref - Jain M, Kushwah KS, Kumar P, Goel AK. Molecular Characterization of Vibrio cholerae O1 Reveals Continuous Evolution of Its New Variants in India. Indian J Microbiol. 2013;53(2):137-141.
Crossref - Goel AK, Jain M, Kumar P, et al. Molecular characterization reveals involvement of altered el tor biotype Vibrio cholerae O1 strains in cholera outbreak at Hyderabad, India. J Microbiol. 2011;49(2):280-284.
Crossref - Borkakoty B, Biswas D, Devi U, Yadav K, Mahanta J. Emergence of classical ctxB genotype 1 and tetracycline resistant strains of Vibrio cholerae O1 El Tor in Assam, India. Trans R Soc Trop Med Hyg. 2012;106(6):382-386.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.