ISSN: 0973-7510
E-ISSN: 2581-690X
Coronaviruses are a group of enveloped viruses with a longer, undivided single-stranded RNA genome, which cause diseases in a variety of animals and humans. In addition to infecting other economically important animals (such as pigs or chickens), six coronaviruses are known to infect human hosts, causing respiratory illness. Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) are highly pathogenic animal coronaviruses that have produced local, regional, and worldwide outbreaks. It is suspected that the current pandemic, caused by a similar coronavirus (SARS-CoVID-19). A new variant of B.1.617 lineage that causes worry about many countries has been identified first in the UK but it makes the worst scenario in India. This includes mutants with immune prolapse E484K and N501Y mutations. Some new variants recently discovered in India like double & triple mutation due to some specific climatic and environmental conditions. Because it creates a viral exodermis and contacts human cells due to mutations in peplomer proteins. The other type of protein is spike protein, are required to bind to receptors in human cells mutations. It can improve the affinity for human receptors and increase the virus, they can cause immune prolapse and reinfection. Moreover, these viruses are capable of adapting and mutating to the new environment. Our immune system is unable to distinguish them from previous infections due to changes in the structure of proteins. The rapid transmission of the COVID-19 around the world causing a severe mortality rate depends on mutation on their spike protein.
COVID-19, SARS-CoV, Variants, Genome, Epidemiology, Mutation
Viruses have always been around mankind and have always been a source of health worry and fatality. These miniscule organisms are always on the list of being the causal organisms of the most fatal and deadly diseases seen in human beings. Even the common cold is caused by a virus that seems to have no cure or vaccine. Diseases like HIV-AIDS, Hepatitis and many more are those diseases whose name is enough to incite fear into their hearts.41 The latest virus to join this army of dangerous viruses is the coronavirus (Covid -19) which, alike to all its family members, exhibit respiratory infection in the host.1
The genetic studies of the virus rather points to the virus being a naturally present animal based virus and the exact know how and when the virus came in contact with humans is not known. There are various theories as to where the virus originated and they seem to coincide with the place where the first recorded case was recorded.1 The city of Wuhan, China in December 2019 saw the first case of respiratory disease whose causal organism was not known but showing symptoms very similar to that of MERS and SARS and within a blink of an eye, the disease spread globally. A meagre 10 months was what took this virus to obtain the momentum to infect an average of 30- 40 million people with alarming number of death rates.42,43
In the beginning, India was well prepared to face the virus due to strict and well put lockdowns and preventive measures. India was so well prepared that it was able to help many countries as well as defend her own borders. The well maintained defence rather cracked with the second wave and India fell in depth of rapid rising cases and endlessly rising piles of death; the advent of delta strains, triple mutant strains don’t seem to make the situation easy for India. India’s miserable situation is getting worse every day for the unsupported infrastructure of healthcare services, premature uplifting of the imposed lockdown and lack of healthy fear towards the ever present virus. Around March 2020, the situation was uncontrolled even after the blockade after the first wave of Indian flu hit. In November, India began to take control of the pandemic situation. Just when things seemed to be under control, in April 2021, India again took possession of Covid’s second wave-19 where the virus was further equipped with various mutations making it more lethal.25
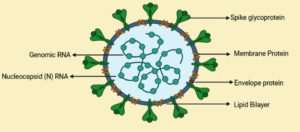
Coronavirus is a type of RNA virus with positive sense. It is composed of a positive-strand RNA genome of the exodermis. Under an electron microscope it has shown that capsid is spirally symmetrical. There are spike-like projections on the surface at 60nm~140nm in diameter, and the crown rises. This is the one of the reasons why it was named coronavirus.1 Since it is similar to SARS-CoV, it will be renamed to SARS-CoV-2 later.2 This disease is caused by SARS-CoV-2 is called coronavirus disease-2019 (year of onset) or COVID-19. The history of coronavirus infection was obtained from the respiratory tract of an adult with a cold. The virus discovered in the mid-1960s was named B814 by Tyrrell and Bynoe. They also conducted studies using human embryonic institutional cultures by inoculating badges from cultures obtained from human volunteers. Since 2003, five new human coronavirus strains have been identified, including the newly discovered Group I coronavirus NL63, NL, SARS, MERS, and New Haven coronavirus, which affect the upper and lower urinary tracts and induces a serious illness. Due to their high mortality, these viral strains are classified as highly contagious viruses that affect mammals, including humans.3 Inoculated with a new viral strain, 229E, to induce a cold and be neutralized with ether. Since the 1930s, various types of coronaviruses that affect animals have been discovered. Among these strains, the strains that mainly infect humans were 229E, OC43, SARS-CoV, NL63, HKU1 and MERS-CoV. COVID-19 has not been previously identified in humans. Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) are two viruses that can cause serious and fatal respiratory illness in humans compared to other strains. Coronaviruses that will affect humans have an envelope and contain positive-stranded RNA. It is a genome based up to 26 to 32 kilobases in size, and so far, it is the largest known genome of an RNA virus4 (Fig. 1).
Fig. 1. Structural characteristic of novel coronavirus showing, Nucleocapsid (N) protein is associated to the single-stranded genomic RNA (ssRNA), that is covered with the aid of using an outer envelope of the primary structural proteins: S glycoprotein (S), membrane protein (M) and envelope protein (E).
Classifications
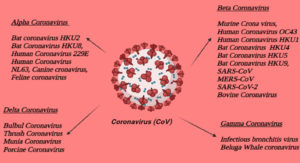
CoVs possess mainly four different genera or subgroup, they are alpha coronavirus, beta coronavirus, gamma coronavirus and delta coronavirus (Fig. 2). It is currently known that all the mentioned CoVs can cause human diseases and belong to the genus Alpha or Beta Coronavirus. Coronaviruses are classified according to the presence of a crown or halo-like shape made up of envelopes containing glycoproteins observed under an electron microscope.44 Only about 7 of these viruses are likely to be severely infected and are listed below.
- OC43 (beta)
- 229E (alpha)
- HKU1 (beta)
- NL63 (alpha)
- MERS-CoV is a beta virus that causes Middle East respiratory syndrome (called MERS)
- SARS beta virus -CoV, can cause severe acute respiratory syndrome (called SARS)
- SARS-CoV-2, which can cause COVID-19.
Molecular Features of Covid-19
The genome of the coronavirus consists of the enzyme complex ORF1ab, which is a 5’non-coded arrangement. S; ORF3a, b, c; E; M; ORF6; ORF7a, b; ORF8; ORF9a, b, c and non-coded regions at the 3′ end and poly A tail 5 (Fig. 3). The largest structural protein, spike protein (protein S) in coronavirus contains N-terminal (S1) and C-terminal (S2) region6. Protein S is responsible for assisting the virus in invading the host cell. The outer subdomain of the SARS-CoV-2, the S1 subunit’s receptor binding domain (RBD) shares roughly 40% amino acid sequence identity with other SARS-related coronaviruses, and the RBD’s essential portion is evolutionary conserved. In addition, the researchers7 discovered one new putative short protein in the SARS-CoV-2 ORF3b domain with four helices “Enzyme II (ACE2)”, which convert Angiotensin in a monocarboxy fold peptase that mediates angiotensin peptide metabolite, and to produce its active form it can cleave angiotensin II. Surprisingly in SARS-CoV-2, RBD’s core domain is highly similar to SARS-CoV, implying that ACE2 is a SARS-CoV-2 receptor2,9 and that the cellular serine protease TMPRSS2 primes SARS-CoV. It suggests that it may be Infection of target cells.8 ACE2 was confirmed as a receptor for SARS-CoV-2 by conducting viral infection experiments in which HeLa cells were expressed or not expressed the protein ACE2. SARS-CoV-2 revealed a high-resolution structure of battlefield ACE2, which is a partial mutation of two S-protein trimers tightly bound to the ACE2 dimer and SARS-CoV-2 S-protein. It suggests that the body may have a closer relationship between RBDs and ACE2. According to another study, SARS-CoV-2 has a higher affinity for the ACE2 receptor than the SARS-CoV S protein, and some monoclonal antibodies against SARS-CoV-S have an effect on SARS-CoV-2 S.10
Fig. 3. Genes encoding structural and non-structural proteins make up the Novel Coronavirus Genome. Nucleocapsid (N), spike (S), membrane (M), and envelope (E) proteins are structural proteins. Each box represents a gene.5
Epidemiological Study
Origin and spread of COVID-19
In December 2019, acute pneumonia of unknown aetiology in adults developed in Wuhan, a province of Hubei and a key transportation centre in China. They also traced live animals and they had found out exposure of the virus in the Huanan wholesale seafood market. The samples from patient collected and sent to labs for etiologic investigations. On December 31, 2019, China alerted the World Health Organization of the outbreak, and the South Korean marine food market was blocked on January 1. The virus was then identified as a coronavirus with >95% homology to bat coronavirus and >70% similarity to SARS-CoV. Environmental samples were also taken from the South Korean seafood market, which were found to be positive. This reveals that the infection originated in that location.11 The number of cases begins to increase exponentially. Some cases are not exposed to the live animal market, so there is a possibility that human-to-human infections are occurring.12 In other countries, Cases of COVID-19 also take place and they don’t have any travel history related to China which indicates that Local “human-to-human transmission” has occurred in these countries.13 In Airport of different country including India, started screening process of people who have symptoms and came from China or other countries for COVID-19 and quarantined them for safety purposes. Soon they noticed asymptomatic people also spreading COVID-19, so they closed all international flights. Though we know that SARS-CoV-2 started from a bat. It is uncertain whether intermediate animals were there, which have moved to close vicinity of humans and have played a pivotal role in spreading of the disease. Pangolins and snakes are currently being suspecting.14
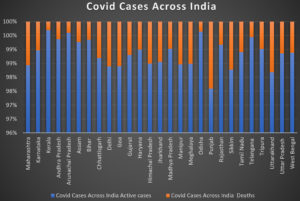
The COVID-19 spread throughout China, the seventh most populous city in China, Wuhan, in early December, but was later exported to more countries. The first verified COVID-19 cases outside of China were reported in Bangkok, Thailand on January 13, 2020.15 As of March 2, 2020, 67 regions outside mainland China have reported 8,565 confirmed COVID-19 cases, causing 132 deaths, and spreading in large numbers of communities in many countries around the world. The organization declared a global pandemic on March 11, 2020.16 Worldwide, the number of confirmed cases has steadily increased. After Asia and Europe, cases in low-income countries have increased sharply.17 Quantifying the pandemic’s exact scope is difficult because all cases must be counted, not just severe and symptomatic cases, but also mild instances.18 However, there has been no consistent worldwide response to this pandemic to date. Each country faces the crisis according to its own capabilities, experiences and assumptions. Therefore, there are different standards for hospitalization, testing, and case estimation, which makes it very much tough to count the number of affected people by this epidemic situation. According to the WHO report, as of May 23, 2021, we have 166,346,635 confirmed cases, 3,449,147 deaths and 1,448,242,899 doses of administered vaccine worldwide (Fig. 4).
Fig. 4. Graphical representation of Covid cases across India (As on 24.05.2021) Data is adapted from, https://www.covid19india.org
Transmission and Symptoms
Virus infection is the entry of the virus into the host cell through a process called endocytosis and also called direct membrane fusion of the integument and host membrane. Once the virus genome enters the host cell, the virus releases uncoated virus particles into the host cell. The 5`cap and 3`polyadenylated tail allow RNA to attach to the host cell ribosome and initiate translation.19. Subsequent cases, when an individual was infected without contact with the viral origin. This conclusion was reached at the beginning of January that Covid-19 was infected as a human being. Some studies have reported that asymptomatic patients can infect others with the virus. Therefore, isolation is the best way to control the Covid-19 virus. Common infections of infectious viruses spread through coughing and sneezing bubbles. Currently, the incubation period of highly infectious coronavirus is known to be 7 days to 2 weeks.20 The virus can infect small water droplets with a diameter of > 5-10 μm. Radio waves generated from the air are generated in the form of water droplets, which can stay in the air for a long time, and water droplet radio waves are generated with a person at a distance of less than 1 m.
Water droplets containing the virus may be dispersed in the air when coughing or sneezing, and it has seen that with the onset of further mutations, the virus is capable of surviving in the suspended form. If you touch or shake hands with a person infected with the virus, the virus may be transmitted to the person. When the virus enters the body via the nose, eyes, or mouth the spikes on the surface can latch on to the surface and undergo structural changes allowing the capsid to fuse with the host membrane. In most of the cases, COVID-19 causes mild symptoms such as a fatigue, fever, and cough (dry). Other mild symptoms include aches and pains, stuffy nose, runny nose, sore throat, and diarrhoea. Some people get infected, but they don’t have any symptoms and don’t feel sick or weak (Table 1).
Table (1):
Symptoms of COVID-19. Data adapted from World Health Organization.
Most common symptoms |
Less common symptoms |
Serious symptoms |
|---|---|---|
• Fever • Dry cough • Tiredness |
• Aches and pains • Sore throat • Diarrhoea • Conjunctivitis • Headache • Loss of taste or smell • A rash on skin, or Discolouration of fingers or toes |
• Difficulty breathing or Shortness of breath • Chest pain or pressure • Loss of speech or movement |
Contact with faeces can transmit some animal coronaviruses, such as the “Feline coronavirus (FCoV).” However, whether this holds true for human coronaviruses is unknown. According to the National Institutes of Health, or NIH, many different types of people are at risk of suffering COVID-19 problems.
Different covid 19 mutant strains of India
All the genetic material of coronaviruses is within the ribonucleic acid or RNA. When the virus infects us, they first attach themselves, then enter our cells, and begin to copy their RNA and spread. If any replication error is a detect during the replication process, the RNA bases will be changed, and these changed bases are called “Mutational changes”. It’s a random process and also the basic mechanism of viruses’ multiplication and spreading. Because of the random ranges, they can make sometime differences in human health and also, they can cause illness. When a virus has a random change, which makes it easier to infect and spreads to the people, that variant will become more common.
On 2021, From the month of January to March, India has received 146 cases of COVID-19 from Maharashtra. Next generation sequencing was performed on nasopharyngeal / oropharyngeal swabs from all symbiotic cases using the Illumina platform21 Through analysis, they searched for 15 SARS-CoV-2 sequences and determined the combination of L452R and E484Q mutations. The scientists attempted to use Vero-CCL-81 cells to isolate the virus from the 15 samples.22 Among the 15 clinical samples, 12 clinical samples showed typical cell rounding, syncytia formation, and cell shedding on the fourth day after infection (PID). As previously mentioned,23 all 12 viruses were isolated, passaged, titrated, sequenced and used for PRNT50 analysis. There are a total of 23 non-synonymous changes in the recovered sequence, with seven conserved non-synonymous mutations in the peak protein (G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H) Variety to the Wuhun-Hu1 sequence. Furthermore, the combination of L452R and E484Q mutations can be seen in various sequences reported to the Global Avian Influenza Data Sharing Initiative by different countries (GISAID).24
Double mutant strain (B.1.617)
This Novel Double mutation, is very much deadlier during the new wave of India and has made it the world’s second worst-hit country. The B.1.617 variant was first turned up in Maharashtra in the month of October. Initially, it was believed to possess two mutations E484Q and L452R, so referred as a double mutant variant.25 The B.1.427 / B.1.429 mutation, which was first detected in California and is associated to enhance transmission, also has a mutation named L452R. The E484Q mutation, which is related to the E484K mutation seen in the P.1 variant, is another B.1.617 mutation. It was first discovered in Brazil, together with the variant B.1.351, also known as the South African variety. The E484K mutation is known as a “escape mutation” because it allows the virus to partially evade immunity conferred by prior infections or immunizations. There are 13 mutations in the genome of B.1.617, which lead to changes in the amino acid sequence of the protein it encodes.26 Three of them are in the virus spike protein, which is very interesting, E484Q, the glutamate-glutamic acid substitution at position 484 causes this variant to bind more to hACE2 (human ACE2 receptor) and has the ability to evade the immune system. Compared to other variants, host performance is better for B.1.61727. L452R substitution at position 452, which is a leucine-arginine replacement, gives the mutant protein a stronger affinity for the ACE2 receptor and lowers immune system recognition. These mutations do not appear to be strain-specific; rather, they appear at the same time.28 P681R, according to William A, this is a substitution at position 681, with proline replacing arginine. Haseltine can increase the variant’s infectivity at the cellular level “by facilitating cleavage of the S precursor protein in an active S1/S2 conformation”.
Table (2):
Novel Coronavirus variant and their spike protein substitution. The data is adopted from Centers for disease control and prevention.
Name |
Spike Protein substitution |
|---|---|
B.1.617 |
Spike: L452R, E484Q, D614G |
B.1.617.1 |
Spike: (T95I), G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H |
B.1.617.2 |
Spike: T19R, (G142D), 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N |
B.1.617.3 |
Spike: T19R, G142D, L452R, E484Q, D614G, P681R, D950N |
Till date, three sub-lines of B.1.617 have been classified. B.1.617.1 (VUI-21APR-01) has been designated as a variant under study by the Department of Public Health of the United Kingdom in April 2021. Two further investigative variants, B.1.617.2 (VUI-21APR-02) and B.1.617.3 (VUI-21APR-03), were discovered at the end of April 2021. B.1.617.3 has the same L452R and E484Q mutations as B.1.617.1, whereas B.1.617.2 lacks the E484Q mutation. In B.1.617.1 and B.1.617.3, the T478K mutation is present.29,30
Triple mutant strain: B.1.618
One SARS-CoV-2 variant (B.1.617) found in India, formerly known as “Triple mutant variant”. This triple mutant takes place when three mutations of a virus combine to make a new variant. In this case, the three mutations are: (i) a deletion and two changes in spike protein (ii) deletion of H146 and Y145, and (iii) mutation in E484K and D614G in spike protein. The B.1.618 is characterised by E484K, a major immune-escape mutation. Two ‘deletions’ in its spike protein, called H146del and Y145del are carried out by this strain. Both these have been associated with immune escape as well.25
This mutant is characterized by the deletion of two amino acids, the peplomer-H146del and Y145del, both of which are linked to immune prolapse, i.e., avoiding antibodies. This variant also has a D614G mutation and an E484K mutation in peplomer, which are associated with increased infectivity and immune prolapse, respectively. We also found that the proportion of SARS-CoV-2 samples from the B.1.618 strain has increased in West Bengal lately and in current situation, it is spreading rapidly across the state.25
Andhra Pradesh Strain
CCMB (Centre for Cellular and Molecular Biology) discovered a new coronavirus variety called N440K. In Visakhapatnam and other parts of the state, the variation is worsening the situation. According to sources, the AP variant was named after Kurnool, where it was initially identified. This novel variant has a shorter incubation period than existing variants, and the disease progresses faster. This new virus has also been found to be 15 times more virulent than previous versions, or even stronger than other Indian strains like B1.617 and B1.618.32
Delta Variant
The variant of Concern of India, known as Delta Plus, was originally mentioned in a Public Health England bulletin on June 11. The variant is a mutant version of B.1.617.2 strain. It is a sub-lineage of the Delta variation that was initially discovered in India and has the K417N spike protein mutation. The mutation K417N has been reported to have “immune evasion” property. On the tip of the coronavirus spike, mutation appears. It binds more tightly to human cells by helping of the virus. It also shows Stronger binding to receptors of lung cells, higher transmissibility. The Mutation K417N was known to reduce the effectiveness of a cocktail of therapeutic monoclonal antibodies so the delta variant also shows “Potential reduction in monoclonal antibody response”.33
India on 23rd June, there have been roughly 40 cases of the variant in Maharashtra, Kerala, and Madhya Pradesh, with “no notable rise in prevalence.” 1st death is reported on 24th June in Madhya Pradesh, India. The Indian government also assured that Covishield and Covaxin are effective against the Delta variant.33
Diagnosis
According to the information posted by CDC and WHO websites, the diagnosis of the new coronavirus assesses the symptoms of patients suffering from a viral infection. Currently, the CoVID-19 test is performed by collecting swabs on the nose and throat that contain the genetic material of the virus. Currently, one of the most efficient diagnostic methods in use is reverse transcription-polymerase chain reaction (RT-PCR), but in a person it can detect a virus only for a while. Also, according to the reports, samples get contaminated during the preparation of laboratory reagents which leads to false positive results. PCR amplification and analysis can be performed simultaneously in a closed system to avoid and minimize false positive results which may occur due to contamination during amplification. I think this is one of the main advantages when it comes to Real Time RT-PCR technology. In its positive sense, coronavirus has a number of molecular targets, including envelope protein (E), transmembrane (M), helicase (Hel), nucleocapsid (N), and glycoprotein spike protein (S). There are various species-specific accessory genes that play a key role in viral replication and can be used for PCR analysis.31
Treatment
Antibody cocktail Casirivimab and Imdevimab
Casirivimab and Imdevimab is an artificial antibody cocktail drug made up of human immunoglobulin G-1 using DNA recombinant technology in the laboratory. These drugs are sold under brand name REGN-COV 2 and are administered in adults and pediatric patients for the treatment of moderate to mild COVID-19 infection. This cocktail drug has been released by Roche and Cipla. Monoclonal antibodies are proteins that mimic the immune system’s ability to fight harmful pathogens (such as viruses). Based on this ability, a monoclonal antibody that specifically targets the SARS-CoV-2 spike protein was designed to prevent the virus from attaching or invading onto human cells.34
2- DG Medicine or Anti Covid drug
In India, the first anti-covid drug known as 2-Deoxy-D-glucose was developed by the Institute of Nuclear Medicine and Allied Sciences, a laboratory of DRDO in collaboration Dr Reddy’s laboratories in Hyderabad. According to the DRDO reports, 2-DG oral medicines will become available in the country after few days as their mass production will begin soon. Since this 2-DG molecule is already aimed towards the treatment of tumors and cancer cells hence, this can be considered as reused drug. 2-DG is a glucose analogue which has an appearance like glucose but it is not glucose. As the virus which grows rapidly inside the body needs glucose for energy production, this particular drug can arrest the virus which inhibits the viral proliferation.35
Dexamethasone or glucocorticoid
Since the outbreak, including in the United States, many doctors have been using these corticosteroids to treat seriously ill COVID-19 patients. It is important for the patients with immune dysregulation or cytokine storm due to some viral infections. In such cases, the lungs and other organs may be damaged, and the overreaction of the immune system can be led to death. These glucocorticoids are effective anti-inflammatory drugs. It is easy to get and inexpensive.36
Remdesivir
In October 2020, the FDA approved antiviral drug Remdesivir was used for the treatment of COVID-19 patients. This drug can be used to treat adults admitted due to COVID-19 and children over the age of 12 weighing 88 pounds or more. Clinical trials have suggested that remdesivir can adequately reduce the recovery time in Covid-19 patients.36
Anticoagulant or blood thinner
In almost all the hospitalized Covid-19 patients’ blood clotting or a high risk of developing blood clotting were observed. These patients were administered with low doses of heparin and enoxaparin in order to prevent blood clots. Due to the high risk of fatality during excessive bleeding, entire volume of anticoagulants was given to balance the risk factor.36
Convalescent plasma
As we have seen from more than a century that blood from recovered patients is used to treat diseases ranging from measles to polio, chickenpox and many others. The convalescent plasma therapy helps the body to produce antibodies to stay healthy in the fight against the coronavirus. Though, this therapy has some conditions to be followed before donating plasma to the Covid-19 patients which includes: donor must be recovered from Covid-19 and is asymptomatic for 14 days, blood compatibility between the donor and acceptor and acceptor must have sufficient antibody titre.37
SARS-CoVID-2 is an unique viral disease caused by a coronavirus that causes severe acute respiratory syndrome. The first and second wave of corona virus is like the “Breaker” of India. As on 23rd april of 2021, the covid cases have crossed the 15.9 million, with 1,85,000 deaths and the second wave is much more devastative compared to the 1st wave.38 Some new variants recently discovered in India like double mutation (B.1.617 ) & triple mutation (B.1.618) due to some specific climatic and environmental conditions. The L452R alteration, which is a leucine-arginine replacement at position 452, gives the mutant protein a greater affinity for the ACE2 receptor and reduces immune system recognition in B.1.61728. B.1.618 is characterised by E484K, a major immune-escape mutation. Two ‘deletions’ in its spike protein, called H146del and Y145del are carried out by this strain.25 Infected people have a chronic cough, fever, sore throat, and myalgia at first, with dyspnea often not appearing until the 14th day of the illness. Dyspnea and hypoxia, together with a persistent temperature, become more prominent in the second stage of the disease, and diarrhoea may become more prominent. In certain cases, a respiratory tract infection develops, requiring ventilation to respiration by the third week of the virus’s infection. COVID-19 diagnostic testing is crucial for detecting the virus, understanding its epidemiology, managing cases, and preventing transmission. Notably, virus detection technology based on nanomaterials can aid in the development of high-sensitivity, simple, scalable, quick, and cost-effective methods for COVID-19.39 Inhibition of viral growth in the early stages of COVID-19 can help to avoid serious consequences later on. In contrast, crucial cases of COVID-19 like anti-inflammation medical aid in conjunction with antiviral agents, as in late stage cytokine release syndrome is the main reason for multi-organ failure and even death.40 Remdesivir appears to decrease the recovery duration of advanced COVID-19 pneumonia, according to clinical evidence. In conclusion, in addition to antiviral medications, the COVID-19 vaccine is the most promising option for ending the present pandemic. COVID-19 patients will benefit from the development of novel antiviral medicines that are specific for SARS-CoV-2.
SARS-CoVID-19 belongs to one of the large families of viruses (coronaviridae), which has a high infectious capability in infecting the host cell, but nowadays, so many vaccines are available worldwide. We can control the spreading, positivity rate by getting vaccinated. In India, there are major chances of getting “Third Coronavirus wave” by November 2021, if we donot maintain the safety guidelines properly. This review sheds light on the current state of COVID-19 and portrays a picture of the current scenario in terms of public health impacts, genome study, and epidemiological study.
The situation is dire and precautionary measures and hygiene standards must be strictly followed if any semblance of normality is to be achieved. It must be understood that we cannot revert back to the prior behaviours and have to be more careful in what we ingest and how we go about in doing our daily routines. We must learn from this incident and try in bettering ourselves in both cultural and scientific aspects.
ACKNOWLEDGMENTS
The authors would like to acknowledge the Management of JSS Academy of Higher Education and Research, Mysuru, Karnataka, for supporting the basic research ideas and also for the resources provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SJ and KMB conceptualized the study. SJ, GB and TSG drafted the manuscript. PR, PRS and KMB helped with the manuscript and discussion.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analysed during this study are included in the manuscript.
- Richman DD, Whitley RJ, Hayden FG. Clinical Virology, 4th ed. Washington: ASM Press. 2016.
Crossref - Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565-574.
Crossref - Kahn JS, McIntosh K. History, and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24(11):S223-S227.
Crossref - Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635-664.
Crossref - Srivastava S, Banu S, Singh P, Sowpati DT, Mishra RK. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J Biosci. 2021;46(1):22.
Crossref - Li W, Wong SK, Li F, et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 2006;80(9):4211-4219.
Crossref - Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221-236.
Crossref - Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.e8.
Crossref - Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
Crossref - Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260-1263.
Crossref - Xinhua. China’s CDC detects a large number of new coronaviruses in the South China seafood market. 2021. http://www.xinhuanet.com/english/2020-01/27/c_138735677.htm. accessed July 14, 2021.
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506.
Crossref - Camilla R, Mirjam S, Peter S, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Eng J Med. 2020; 382(10):970-971.
Crossref - Singhal T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediatr. 2020;87(10223):281-286.
Crossref - Sharun K, Sircar S, Malik YS, Singh RK, Dhama K. How close is SARS-CoV-2 to canine and feline coronaviruses?. J Small Anim Pract. 2020;61(8):523-526.
Crossref - Hsu LY, Chia PY, Lim JF. The Novel coronavirus (SARS-CoV-2) epidemic. Ann Acad Med Singap. 2020;49(3):105-107.
Crossref - World Health Organization Coronavirus Disease 2019 Situation Report. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200326-sitrep-66-covid-19.pdf. accessed 26 March, 2020.
- Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of covid-19-Studies needed. N Engl J Med. 2020;382(13):1194-1196.
Crossref - Graham S, Zmora P, Gierer S, Heurich A, Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605-614.
Crossref - Cascella M, Rajnik M, Aleem A, Dulebohn SC, SC RD Napoli. Features, evaluation, and treatment coronavirus (CoVID-19), Stat Pearls Publishing”. Treasure Island (FL). 2020.
- Yadav PD, Nyayanit DA, Shete AM, et al. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis. 2019;10(1):23-33.
Crossref - Sarkale P, Patil S, Yadav PD, et al. First isolation of SARS-CoV-2 from clinical samples in India. Indian J Med Res. 2020;151(2):244-250.
Crossref - Deshpande GR, Sapkal GN, Tilekar BN, et al. Neutralizing antibody responses to SARS-CoV-2 in COVID-19 patients. Indian J Med Res. 2020;152(1):82-87.
Crossref - Yadav PD, Sapkal GN, Abraham P, et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin Infect Dis. 2020.
Crossref - Sahoo JP, Mishra AP, Samal KC. Triple Mutant Bengal Strain (B.1.618) of Coronavirus and the Worst COVID Outbreak in India. Biotica Research Today. 2021;3(4):261-265.
- Expert reaction to cases of variant B.1.617 (the ‘Indian variant’) being investigated in the UK | Science Media Centre. Science Media Centre. Accessed 26 April, 2021.
- Hoffmann M, Hofmann-Winkler H, Kruger N, et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3):109415.
Crossref - Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Reports Medicine. 2021;2(4):100255.
Crossref - SARS-CoV-2 Variant Classifications and Definitions. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html , accessed 12 May, 2021.
- Di Giacomo S, Mercatelli D, Rakhimov A, Giorgi FM. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J Med Virol. 2021;93(9):5638-5643.
Crossref - Tang YW, Schmitz JE, Persing DH, Stratton C. The Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges. J Clin Microbiol. 2020;58(6):e00512-20.
Crossref - CCMB Refutes ‘N440K COVID Strain 15 Times More Lethal’; Says ‘Andhra Strain Diminishing’. Republic World. https://www.republicworld.com/india-news/general-news/ccmb-refutes-n440k-covid-strain-15-times-more-lethal-says-andhra-strain-diminishing.html, accessed 12 May, 2021.
- Delta Variant: What We Know About the Science. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/variants/delta-variant.html, accessed 30 August, 2021
- Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19. U.S. Food and Drug Administration. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19, accessed 12 May, 2021.
- DCGI approves anti-COVID drug developed by DRDO for emergency use. PIB Delhi. https://pib.gov.in/PressReleasePage.aspx?PRID=1717007 , accessed 12 May, 2021.
- COVID-19 Treatment Guidelines. National Institutes of Health. https://www.covid19treatmentguidelines.nih.gov/, accessed 13 june, 2021.
- Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20(4):398-400.
Crossref - Jain VK, Iyengar KP, Vaishya R. Differences between First wave and Second wave of COVID-19 in India. Diabetes Metab Syndr. 2021:15(3):1047-1048.
Crossref - Kevadiya BD, Machhi J, Herskovitz J, et al. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021:20(5):593-605
Crossref - Chen P-L, Lee N-Y, Cia C-T, Ko W-C, Hsueh P-R. A Review of Treatment of Coronavirus Disease 2019 (COVID-19): Therapeutic Repurposing and Unmet Clinical Needs. Front Pharmacol. 2020:11:584956.
Crossref - Snowden FM. Emerging and reemerging diseases: a historical perspective. Immunol Rev. 2008:225(1):9-26.
Crossref - Keni R, Alexander A, Nayak PG, Mudgal J, Nandakumar K. COVID-19: Emergence, Spread, Possible Treatments, and Global Burden. Front Public Health. 2020;8:216.
Crossref - Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181-192.
Crossref - Tyrrell DAJ, Myint SH. Coronavirus, Medical Microbiology. Univer sity of Texas Medical Branch at Galveston. 4th ed. chapter 60. 1996.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.