ISSN: 0973-7510
E-ISSN: 2581-690X
The three human Enterovirus serotypes D-68, D-70, and A-71, are common pathogens that are transmitted by fecal-oral and aerosol routes. These positive RNA viruses were known to exhibit high levels of genetic diversity and variability. Currently, no vaccines are available to protect humans from these three serotypes. Therefore, efforts are needed for the development of a vaccine directed against heterologous viruses. In our study, an immunoinformatics approach is used to identify T- and B-cell epitopes that may help for the generation of a universal vaccine against EV-D70, EV-A71, and EV-D68. B and T cell epitopes were selected based on their length. As a result, 5 B cell epitopes and 18 T cell epitopes were predicted. Our B cell epitope prediction results showed that there are a number of linear regions. Position 150-170 was found to be the most immunogenic for the different strains. Regarding the epitopes of the T lymphocytes, the result of the interactions shows that 95% of the predicted epitopes are common between the 3 sequences and the 5 methods used. These results demonstrate the great immunogenic potential of these sequences and their capacities to trigger immune reactions in people with different HLA alleles. The “VFYDGFAGF” epitope is the most important and most immunogenic for triggering an immune response. Our study results allowed us to identify epitopes to be used in the development of cross-protection vaccines against the three Enterovirus serotypes. However, in vivo and in vitro studies are needed to assess the potential of the epitopes predicted by our study.
Enterovirus, Epitopes, VP1 structural protein, Computational prediction
Human enteroviruses (EV) EV-D68, EV-D70, and EV-A71 belong to the genus Enterovirus of Picornaviridae.1 They are highly resistant to external environmental conditions. Their transmission mode is mainly faecal-oral, although they can also be transmitted by aerosols.2 There does not appear to be a relationship between a given genotype and a particular geographic area.3 Enteroviruses (EVs) are common infectious agents and they currently comprise 108 stereotypes. They are non-enveloped, single-stranded, positive-polarity RNA viruses, characterized by a genome size of approximately 7500 nt that codes for 11 viral proteins. Their capsid consists of 60 identical protomers, or capsomers, each containing 4 structural viral proteins (VP) sach us: VP1 (34 kDa), VP2 (30 kDa), VP3 (26 kDa), and VP4 (7 kDa). VP1, VP2, and VP3 form a wide depression of 15 Å on the surface of the virus, called a canyon, which is thought to be the interaction site of the virus and its receptor.4,5
The VP1 protein encoded by the 1D protein gene is largely exteriorized on the surface of the virion and it bears several antigenic epitopes located mainly in the peptide loops that connect the beta-sheets.6 The VP1 protein of EV-D68, EV-D 70, and EV-A71 is involved in attachment of the virus to the host cell,7 not only as a major structure of the canyon site but also due to its antigenicity, as it bears a set of epitopes involved in specific recognition of these viruses.8
In light of the pathogenicity of EVs, the currently available therapeutic means are very limited or non-existent in clinical practice.9 The development of new molecules and/or antiviral strategies is hence a necessity and should lead to greater knowledge of the infection cycle of the target cell, in particular, the processes of attachment and entry of the virus into the cell.10
Given the technological potential of subunit vaccines compared to conventional whole virus vaccines, we investigated the benefit of using the VP1 subunit of enteroviruses is sufficient to obtain adequate protective immunity. Due to the antigenic importance of the three-dimensional structure of viral capsids in which all the capsid proteins function in concert, the application of VP1 subunit vaccines for enteroviruses has always been reluctant, despite the fact that the agglutination of many epitopes on VP1 independent of other capsid proteins has been widely demonstrated.11
In our research, we used an immunoinformatics approach based on computational approaches for predictive vaccines in order to select common epitopes for the three serotypes to evaluate the conservation of the peptide sequences of EV-D68, EV-D70, and EV-A71 at the level of the VP1 protein using reference sequences from the NCBI genome bank. Subsequently, linear epitopes of B lymphocytes and T lymphocytes were predicted and their accessible surface identified as well as their ability to generate an immune response.
The analysis of the results obtained will be used to consider a therapeutic treatment that can contribute to a reduction of infections caused by these three enteroviruses.
In this work, we used modeling tools in order to study and determine the potential epitopes of VP1 that can be used for the purpose of vaccination against EV-D68, EV-D70, and EV-A71.
Data set
The following amino acid sequences of VP1 of serotypes A, B, and C were obtained from the National Biotechnology Information Centre (NCBI) database: 183 aminos acids of VP1 of EV-D68 (AYG85340.1), 310 aminos acids of VP1 of EV-D70 (BAA08157.1), and 299 aminos acids of VP1 of EV-A71 (ACS12928.1).
Sequences
Different genotypic protein sequences of enterovirus protein VP1 were obtained from the National Biotechnology Information Centre (NCBI). Complete sequences of all subtypes of different genotypes were selected. Sequences containing only partial sequences of genotypes were excluded.
After obtaining the NCBI protein sequences, multiple sequence alignments were performed using Muscle MegaX software. The default settings were used on the interface. The highest numbers of similar and identical amino acids without gaps covering the protein sequence were selected as conserved areas. These conserved areas were used to predict B-cell linear epitopes, surface-accessible epitopes, and antigenic sites.
Protein structure Prediction and consensus sequence validation
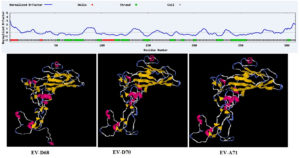
Subsequently, the reference sequences for VP1 of genotypes A, B, and C were submitted to the Swiss-Modeler server12 for the prediction of the homology degree of the 3D structure. Three-dimensional proteomic structures with a greater than 80% identity and that had a statistically significant homology (e-value0.001) were selected.
The sequences for which these conditions do not apply were modeled with the I-TASSER13 server in order to predict the 3D structure of the protein using an ab initio computational approach that can also predict their biological function. The consensus sequence was uploaded to the I-TASSER server for thread structure prediction (http://zhanglab.ccmb.med.umich.edu/I-TASSER). The planned structure was refined by ModRefiner.14 The quality of the structure was confirmed by the Ramachandran plot.15
Epitopes prediction
The B- and T-cell epitopes prediction of was made using methods that differ from the common B- or T-cell epitopes, among all of the methods used, and were determined via
- For “T”-cell epitopes
The tools offered in IEDB were used. Before predicting the epitopes, each epitope was submitted to analysis by the following algorithms: BIMAS,16 SYFPEITHII,17 and NetMHC.18
Epitope prediction tools were used to check if the epitopes can be presented on the surface of the cells, The T-cell epitope prediction tools were used to predict the potential epitopes of the EV-VP1 protein: MHC-I, MHC-NP, NetCTLpan 1.1, RANKPEP,19 and NetMHCpan 3.0 treatment predictions. Since HLA-A0201 was the most common CMH allele and most of the MHC-I epitopes were monopeptides, the allele and monopeptide HLA-A0201 were selected. The other parameters of each prediction tool were set by default.
The purpose of the MHC-I IEDB treatment prediction server is to identify CMH-I ligands. This website provides a tool for the prediction of protein-treated peptides naturally presented by MHC Class I molecules. The MHC-I linkage forecast was established on March 30, 2019, using the consensus of the IEDB analysis resource, which combines the forecasts of ANN, aka NetMHC (3.4), SMM, and Comblib (http://tools.immuneepitope.org/processing/).
The MHC-NP server20 is devoloped for the prediction of peptides naturally presented by CMH molecules. This website uses data from CMH elution experiments to assess the probability of a given peptide being treated naturally and its binding to a given CMH molecule (http://tools.immuneepitope.org/mhcnp/). The NetCTLpan 1.1 server is designed to predict the epitopes of cytotoxic lymphocytes (CTL) in protein sequences. This website integrates the predicted link to the MHC-I peptide, the cleavage of the C-terminal proteasomal cleavage site using artificial neural networks (ANN),21 and the transport efficiency of the TAP (protein), which was predicted using the weight matrix (http://www.cbs.dtu.dk/services/NetCTLpan /).
The purpose of the RANKPEP server is the prediction of the epitopes of CMH Class I and Class II molecules based on protein sequences. This method predicts epitopes using position-specific score matrices (PSSM) (http://imed.med.ucm.es/Tools/rankpep.html). The NetMHCpan3.0 server is designed to predict the peptide-MHC binding capacity of Class I.
- For the “B”-cell epitopes prediction:
Four different methods were used to predict the B cell epitopes: HMM (Hidden Markov Model), ANN (artificial neural network),21 and deep learning methods.22 We defined the threshold of positive epitopes if they are equal or bigger than 1.0 in the Immune server package Epitope DataBases (IEDB),23 The epitopes were validated by the method of Chou and Fasman beta turn.
VP1 3D models and alignments
Although that in PDB database, there is numerous crystallized structures from the complete virus. The screening condition to select the protein were fall done due to the Bigger Resolution which is often bigger than 3 Å. In addition, as we would like to include the mutation observed in the sequences recently published. We used the ab initio approach to model 3D structures and to optimize structures via ModRefined (Fig. 1). The published structure in PDB were almost similar to our modeling results with a few modifications due to the mutations recently revealed.
VP1 “T”-cell epitopes
In a pre-selected environment, the IEDB recommends MHC-NP, NetCTLpan 1.1, RANKPEP, and the NetMHCpan 3.0 server to predict powerful epitopes of the sequence. In order to improve the accuracy of epitope prediction, epitopes predicted by at least four tools have been selected. (the full prediction of epitopes is included in the Supplementary Data). The analyses showed that HLA-A*01:01 has greater predictive value for the 3 sequences.
Table (1):
List of 18 predicted epitopes common for T lymphocytes of the three serotypes (EV-D68, EV-D70, EV-A71).
Allele |
Start |
End |
Peptide Length |
Peptide |
Proteasome score |
TAP score |
MHC score |
Processing score |
Total score |
MHC IC50 (nM) |
|---|---|---|---|---|---|---|---|---|---|---|
HLA-A*01:01 |
266 |
274 |
9 |
YTNILNNNY |
1.39 |
1.15 |
-1.41 |
2.54 |
1.13 |
25.5 |
HLA-A*23:01 |
175 |
183 |
9 |
VFYDGFAGF |
1.31 |
1.30 |
-1.42 |
2.62 |
1.19 |
28.0 |
HLA-A*02:06 |
174 |
183 |
10 |
SVFYDGFAGF |
1.31 |
1.21 |
-1.88 |
2.52 |
0.65 |
75.3 |
HLA-A*01:01 |
266 |
275 |
10 |
YTNILNNNYY |
1.20 |
1.15 |
-1.26 |
2.35 |
1.08 |
18.3 |
HLA-A*24:02 |
234 |
242 |
9 |
SYQPVQYTL |
1.60 |
0.60 |
-1.16 |
2.20 |
1.05 |
14.3 |
HLA-A*24:02 |
197 |
205 |
9 |
AYANFYDGF |
1.35 |
1.31 |
-1.61 |
2.66 |
1.04 |
41.1 |
HLA-A*24:02 |
196 |
205 |
10 |
SAYANFYDGF |
1.35 |
1.29 |
-1.88 |
2.64 |
0.76 |
75.2 |
HLA-A*24:02 |
132 |
140 |
9 |
LFSSSNVSF |
1.55 |
1.23 |
-2.15 |
2.78 |
0.63 |
142.4 |
HLA-A*02:06 |
26 |
34 |
9 |
GVIPSLNAV |
1.19 |
0.17 |
-0.78 |
1.36 |
0.58 |
6 |
HLA-A*11:01 |
193 |
202 |
10 |
SINSAYANFY |
1.33 |
1.32 |
-2.17 |
2.66 |
0.49 |
147.2 |
HLA-A*24:02 |
131 |
140 |
10 |
RLFSSSNVSF |
1.55 |
1.28 |
-2.39 |
2.84 |
0.45 |
243 |
HLA-A*24:02 |
111 |
120 |
10 |
KMELFTYLRF |
1.37 |
1.19 |
-2.13 |
2.56 |
0.44 |
134.1 |
HLA-A*01:01 |
192 |
202 |
11 |
MSINSAYANFY |
1.33 |
1.29 |
-2.21 |
2.62 |
0.42 |
160.8 |
HLA-A*02:01 |
117 |
130 |
14 |
YLRFDTEITIVPTL |
1.74 |
0.44 |
-1.79 |
2.19 |
0.40 |
61.5 |
HLA-A*11:01 |
72 |
81 |
10 |
ALVCMRSFEY |
1.30 |
1.40 |
-2.36 |
2.70 |
0.34 |
228.9 |
HLA-A*11:01 |
262 |
275 |
14 |
RTMPYTNILNNNYY |
1.20 |
1.33 |
-2.20 |
2.53 |
0.33 |
157 |
HLA-A*02:06 |
262 |
270 |
9 |
RTMPYTNIL |
1.73 |
0.48 |
-1.87 |
2.21 |
0.33 |
74.8 |
HLA-A*11:01 |
73 |
81 |
9 |
LVCMRSFEY |
1.30 |
1.40 |
-2.39 |
2.71 |
0.31 |
246.3 |
HLA-A*26:01 |
222 |
235 |
14 |
NTMGNLCLRVVNSY |
1.28 |
1.37 |
-2.40 |
2.66 |
0.25 |
252.6 |
HLA-A*02:01 |
117 |
126 |
10 |
YLRFDTEITI |
1.32 |
0.26 |
-1.33 |
1.58 |
0.25 |
21.3 |
The result of the interactions shows that 95% of the predicted epitopes are common between the 3 sequences and the 5 methods used. These results thus demonstrate the great immunogenic potential of these sequences and their capacities to trigger immune reactions in people with different alleles However, it should be pointed out that the immunogenic rate differs between the sequences and the epitopes. The table above presents the list of the 18 epitopes predicted for VP1 T lymphocytes.
Epitope “VFYDGFAGF” is the most important and immunogenic one, with a percentage of 90% and an IC of 28 nM to trigger an immune reaction. However, the other epitopes require more than 100 nM. This counts better for several reasons including TAP and MHC. Other shared epitopes of the same importance are shown in the Supplementary data. “SSN” epitopes get a response on 225 predicted positions. An advanced understanding of antigen recognition at the molecular level enables the development of an epitope vaccines. The approach to building an epitope vaccine is based on the identification of B-cell epitopes and T-cell epitopes. These epitopes are immunodominant and have the capacity to induce a specific immune response.
VP1 “B”- cell epitopes
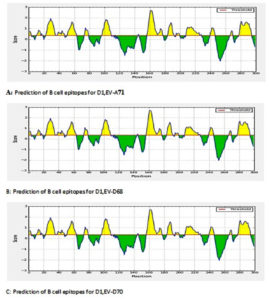
For B epitopes, linear epitopes were identified by the HMM and ANN methods, which showed that there are a number of linear regions. The position 150-170 was found to be the most immunogenic for the different strains (Fig. 2)
Enteroviruses are responsible for several diseases that adversely impact public health, notably neurological manifestations: lymphocytic meningitis and flaccid paralysis (EV-D70 and EV-A71)24, 25, skin and mucosal membrane manifestations: hemorrhagic conjunctivitis, MPB syndrome (EV-A71)26, and rhinopharyngitis and respiratory infections: bronchiolitis (EV-D68).27
The EV capsid VP1 protein is one of four structural proteins of EV. Since the VP1 protein is highly exposed and has been reported to play an important role in viral pathogenesis and virulence,28 and its antigenic homology among many different EV serotypes has been well documented.29 The N termini of most EV VP1 proteins contain highly conserved immunogenic regions that are recognized by sera from most EV-infected patients.30 This results suggests the potential use of the EV VP1 protein as a target to raise pan-EV mAbs (monoclonal antibodies) for the detection of a broad spectrum of EVs.
To make the molecule immunogenic, B-cell epitopes may be linked to T-cell epitopes. Advances in bioinformatics have had significant impact in immunology sector, which allowed the rapid development of computer immunology also called immunoinformatics
Immunoinformatics research considerably assists with mapping T and B epitopes of pathogens. Using various bioinformatics tools, immunologists are able to analyze potential sequences and their linking sites in a short time frame. This can lead to the development of new efficient vaccines. In this research, various bioinformatics algorithms to develop an effective multi-epitope vaccine were used. In addition to the existence of 68 viral serotypes, the antigenic properties change even within a given strain.
The conserved areas were analyzed for the identification of B-cell epitopes, antigenicity of the epitopes, accessibility to the surface, conservation of the epitopes, and hydrophilia in order to select the final epitopes of B cells. In addition, the conserved regions were analyzed and T-cell epitopes were also identified. The epitopes predicted from the conserved areas exhibited all of the properties described above. The Immune Epitope Database (IEDB) (http://tools.immuneepitope.org/bcell/) was used to predict antigenic sites. Kolaskar and Tongaonkar antigenicity methods were used for to predict antigenic sites with a default threshold value of 1.0. The B-cell epitopes were predicted using the consensus sequence. The prediction of T-cell epitopes were made using alleles for haplotypes A18, A31, A19, A14, A20, A11, A13, and A10 in which the Class I HMC genes were expressed. Selection of the epitopes was carried out and the duplicate epitopes were removed. The selection of B and T cells epitopes were based on length. As a result, 5 B lymphocyte and 18 T lymphocyte epitopes were predicted.
Novelty of this work lies in the evaluation of the possibility to synthesize a common vaccine for the three serotypes, although other work may have highlighted vaccines for EV-A71 or EV-D70.
Synthetic peptide vaccines designed to achieve specific immune responses against pathogens have several advantages. First, they are relatively simple and cost-effective to produce, and secondly, a large variety of peptide vaccines can be synthesized.31 While this area of research is not yet well established, a lot of work is currently being carried on synthetic peptide vaccines.
In the same line with our results, the study conducted by Park et al32 regarding the development of a synthetic vaccine for several EV-D70 strains showed that E peptide-induced antibodies should protect against most EV-D70 strains, and their results highlight the potential of the E peptide as a synthetic peptide vaccine against EV-D70, while the results from the study of Chen et al33 demonstrated that an N-terminal fragment (aa 1–138) of EV-D70 VP1, expressed by a bacterial expression system, induces more neutralizing antibodies than a C-terminal fragment (aa 141-310). These studies corroborate our results. Chen et al33 showed that the N- and C-terminal fragments have independent antigenic neutralization sites, which is consistent with our results, provided that the B epitopes that we predicted are located in the same regions. Therefore, EV-D70 VP1 appears to have several independent neutralizing epitopes. Particularly important is the observation that the E-peptide region in the N-terminus of VP1 is a powerful B epitope that can induce the production of neutralizing antibodies against EV-D70. Thus, it could be a possible target for the development of an effective vaccine against EV-D70 peptides.32
Another study, by,34 described two subunits of EV-A71 vaccines, one as a DNA vaccine and the other as a recombinant protein vaccine that induced a neutralizing antibody response in both ICN and BALB/c mice. These results provide evidence that VP1 of EV71 contains neutralizing epitopes independent of other viral capsid proteins and lead the way for the potential use of VP1 as a basic antigen for the development of EV-A71 subunit vaccines.
The special form of the antigen is generally considered to be a better independent activator of T and B lymphocytes35,36 compared to the soluble antigen. Nevertheless, if these mechanisms works in EV-A71, immunity should be studied empirically.34 These results were verified by our study, which led to the prediction of B epitopes but also 18 T epitopes that are immunodominant and that have the ability to induce a specific immune response.
However, for EV-D68, the work by37 has shown that, since the first virus isolation, mutations have been observed in the BC and DE loops of the VP1 sequence, which corresponds to the region of the protein on the viral surface linked to antigenic epitopes, therefore indicating that unique sequence variations in these loops may cause a change in antigenicity, although there is little data on serotype vaccination in the current literature. Therefore, our study adds significantly in this regard.
This in silico study represents a major step forward in the development of a common vaccine against viruses that present a major risk to public health. However, the feasibility of such vaccine development must be verified by in vivo studies.
As a better way to use the results of this study, this work can be considered as a basis for in vivo studies.
ACKNOWLEDGMENTS
Authors would like to thank the Ministry of High Education of Morocco, the University Hassan II of Casablanca and Faculty of sciences and techniques Mohammedia for their support. We would also like to thank all the staff of Laboratory of Virology, Mocrobiology, Quality and Biotechnologies / Ecotoxicology and Biodiversity and the team of Virology for their technical assistance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
BR, HN, BH, MME conceptulized the project. BR, KY, HN proposed the methodology. BR, KY, BH, MME validated the results. BR wrote the manuscript. HN reviewed the manuscript. All authors read and approved the manuscript for the final publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Tapparel C, Siegrist F, Petty TJ, Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282-293.
Crossref - AFSSA (Agence Francaise de Securite Sanitaire des Aliments). Actualisation de l’exposition alimentaire au chlordecone de la population antillaise, evaluation de l’impact de mesures de maitrises des risques. 2007. https://www.anses.fr/fr/system/files/RCCP-Ra-ChlAQR2007.pdf
- Bailly C, El-Maarouf-Bouteau H, Corbineau F. From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. Comptes Rendus Biologies. 2008;331(10):806-814.
Crossref - Grant RA, Hiremath CN, Filman DJ, Syed R, Andries K, Hogle JM. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr Biol. 1994;4(9):784-797.
Crossref - Colston E, Racaniello VR. Soluble receptor resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. The EMBO Journal. 1994;13(24):5855-5862.
Crossref - Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human Enterovirus: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73(3):1941-1948.
Crossref - Wang M, Larsen MV, Nielsen M, et al. HLA class I binding 9mer peptides from influenza A virus induce CD4 T cell responses. PLoS One. 2010;5(5):e10533.
Crossref - Langeveld JP, Casal JI, Vela C, et al. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J Virol. 1993;67(2):765-772.
Crossref - Barnard DL. Current status of anti-picornavirus therapies. Current Pharmaceutical Design. 2006;12(11):1379-1390.
Crossref - Nilsson EC, Jamshidi F, Johansson SMC, Oberste MS, Arnberg N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol. 2008;82(6):3061-3068.
Crossref - Xie Q, He X, Yang F, et al. Analysis of the genome sequence and prediction of B-cell epitopes of the envelope protein of Middle East respiratory syndrome-coronavirus. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB). 2018;15(4):1344-1350.
Crossref - Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research. 2018;46(W1):W296-W303.
Crossref - Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research. 2015;43(W1):W174-W181.
Crossref - Xu D, Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophysical Journal. 2011;101(10):2525-2534.
Crossref - Ramachandran GT, Sasisekharan V. Conformation of polypeptides and proteins. In Advances in protein chemistry, Academic Press. 1968;23:283-437.
Crossref - Mears LA, Afiff S. A new look at the BIMAS program and rice production. Bulletin of Indonesian Economic Studies. 1968;4(10):29-47.
Crossref - Rammensee H-G, Bachmann J, Emmerich NPN, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213-219.
Crossref - Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Research. 2008;36(Suppl_2):W509-W512.
Crossref - Reche PA, Glutting JP, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56(6):405-419.
Crossref - Giguere S, Drouin A, Lacoste A, Marchand M, Corbeil J, Laviolette F. MHC-NP: Predicting peptides naturally processed by the MHC. J Immunol Methods. 2013;400-401:30-36.
Crossref - Zurada JM. Introduction to artificial neural systems. St. Paul: West publishing company. 1992;8.
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444.
Crossref - Kim Y, Ponomarenko J, Zhu Z, et al. Immune epitope database analysis resource. Nucleic Acids Research. 2012;40(W1):W525-W530.
Crossref - Wanjiru MA. Isolation and characterization of human Enterovirus from stored samples of children with acute respiratory infections attending Kenyatta National Hospital, Nairobi. Diss. 2014.
- Huang SW, Kiang D, Smith DJ, Wang JR. Evolution of re-emergent virus and its impact on enterovirus 71 epidemics. Exp Biol Med. 2011;236(8):899-908.
Crossref - Farias AA, Mojsiejczuk LN, Pisano MB, et al. Environmental Surveillance of Enterovirus in Central Argentina: First detection and evolutionary analyses of E14. Food Environ Virol. 2018;10(1):121-126.
Crossref - Ravishankar S, Chapin K, Alexander-Scott N, et al. Enterovirus D68 and Panton-Valentine Leukocidin-Positive Staphylococcus aureus Respiratory Coinfection with Fatal Outcome. Pediatr Dev Pathol. 2016;19(1):80-85.
Crossref - Tan SH, Ong KC, Perera D, Wong KT. A monoclonal antibody to ameliorate central nervous system infection and improve survival in a murine model of human Enterovirus-A71 encephalomyelitis. Antiviral research. 2016;132:196-203.
Crossref - Mao Q, Wang Y, Shao J, et al. The compatibility of inactivated-Enterovirus 71 vaccination with Coxsackievirus A16 and Poliovirus immunizations in humans and animals. Human Vaccines & Immunotherapeutics. 2015;11(11):2723-2733.
Crossref - Zhang J, Huang H, Xu L, Lou C, Pan M. Screening and Identification of Linear B Cell Epitopes Within the Nonstructural Proteins of Enterovirus 71. Viral immunol. 2019;32(2):84-88.
Crossref - Wang HY, Tsao KC, Hsieh CH, et al. Inferring nonneutral evolution from contrasting patterns of polymorphisms and divergences in different protein coding regions of enterovirus 71 circulating in Taiwan during 1998-2003. BMC Evol Biol. 2010;10:294.
Crossref - Park KB, Lim BK, Ye MB, Chung SY, Nam JH. A peptide vaccine based on a B-cell epitope on the VP1 protein of enterovirus 70 induces a strong antibody response. Acta Virol. 2012;56(4):337-342.
Crossref - Chen D, Duggan C, Texada DE, Reden TB, Kooragayala LM, Langford MP. Immunogenicity of enterovirus 70 capsid protein VP1 and its non-overlapping N- and C-terminal fragments. Antiviral Res. 2005;66(2-3):111-117.
Crossref - Wu CN, Lin YC, Fann C, Liao NS, Shih SR, Ho MS. Protection against lethal enterovirus 71 infection in newborn mice by passive immunization with subunit VP1 vaccines and inactivated virus. Vaccine. 2001;20(5-6):895-904.
Crossref - Fehr T, Skrastina D, Pumpens P, Zinkernagel RM. T-cell-independent type I antibody response against B-cell epitopes expressed repetitively on recombinant virus particles. Proc Natl Acad Sci USA. 1998;95(16):9477-9481.
Crossref - Morein B, Simons K. Subunit vaccines against enveloped viruses: virosomes, micelles and other protein complexes. Vaccine. 1985;3(2):83-93.
Crossref - Patel MC, Wang W, Pletneva LM, et al. Enterovirus D-68 infection, prophylaxis, and vaccination in a novel permissive animal model, the cotton rat (Sigmodon hispidus). PLoS One. 2016;11(11):e0166336.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.