ISSN: 0973-7510
E-ISSN: 2581-690X
The prevalence of multidrug-resistant gram-negative bacilli has increased worldwide. Critical care areas of most hospitals use carbapenem antibiotics for the empirical treatment of gram-negative bacterial (GNB) infections. In the last decade, there have been reports of the detection of carbapenem-resistant Enterobacterales (CRE). This rise in the spread of CRE presents a great challenge in the treatment of GNB infections and poses a serious threat to global health. To detect the burden of CRE and to characterize CRE, we used three phenotypic methods for the detection of carbapenemase enzymes. Using conventional aerobic bacterial culture methods, 150 Enterobacterales strains were isolated from various clinical samples. Identification of CRE was performed using multiple phenotypic detection methods, such as the Kirby Bauer disc diffusion method for meropenem (10 mcg) using the CLSI 2021 interpretation for meropenem, modified Hodge test (MHT), Carba NP test, and modified carbapenem inactivation method (mCIM) test. A total of 150 Enterobacterales strains were isolated over a period of 1 year. Among these, 66/150 (44%), 63/150 (43%), 64/150 (43%), and 65/150 (43%) were identified as CRE using the Kirby Bauer disc diffusion method, MHT, mCIM test, and Carba NP test, respectively. The sensitivity and specificity of MHT, mCIM, and Carba NP tests within 95% CI were 93.94%/100%, 96.97%/100%, and 98.48%/100%, respectively. The positive and negative predictive values of MHT, mCIM, and Carba NP tests were 100%/95.45%, 100%/97.67%, and 100%/98.82%, respectively. The accuracies of the MHT, mCIM, and Carba NP tests were 97.33%, 98.67%, and 99.33% respectively indicating a high burden of carbapenem resistance in Enterobacterales. Therefore, given the current statistics of carbapenem resistance, use of carbapenem as empiric treatment in the intensive care units of hospitals may not be beneficial. Identification of carbapenem resistance can help in the initiation of appropriate antimicrobial therapy. This study compares the accuracy and efficiency of Carba NP, mCIM, and MHT in detecting carbapenem-resistant Enterobacterales.
Carba NP, Modified Hodge Test, Modified Carbapenem Inactivation Method, Multidrug Resistant Gram Negative Bacilli, Carbapenem Resistance, Enterobacterales
Infections caused by multidrug-resistant (MDR) Enterobacterales are a threat to global health owing to the high mortality rates caused by these strains. Carbapenem-resistant Enterobacterales (CRE) rank among the top tier of antibiotic-resistant “priority pathogens” along with carbapenem-resistant Acinetobacter baumanii (CRAB) and carbapenem-resistant Pseudomonas aeruginosa (CRPA). The Indian Council of Medical Research (ICMR) has estimated the multidrug resistance in Escherichia coli and Klebsiella pneumoniae to be 30% and 50% respectively.1
Treatment options available for MDR Enterobacterales are limited, with carbapenems being the drugs of choice for the treatment of infections caused by Enterobacterales that produce extended-spectrum β-lactamase (ESBL). Carbapenems are pivotal in the prophylaxis and treatment of infections in transplant surgery, intensive care units, and during other surgical procedures. Carbapenems are β-lactam antimicrobials that have a high level of activity against ESBL-producing bacteria and can be administered intravenously. Carbapenems are the most dependable and last-resort antibiotic class for treating bacterial infections.2,3
There have been numerous reports of carbapenemase-producing Enterobacterales across the world.4,5 Plasmids and transposons have been shown to be responsible for the transfer of carbapenem resistance across bacterial species such as E. coli, Salmonella spp., Citrobacter freundii, and Enterobacter cloacae. The presence of structural mutations in β-lactamases such as ESBL, AmpC, and carbapenemase, are the major mechanisms by which resistance is conferred in CRE. Among the carbapenemases, Klebsiella pneumoniae carbapenemase (KPC, Class A), imipenemase (IMP, Class B), Verona integrin-encoded MBL (VIM, Class B), New Delhi metallobetalactamase (NDM, Class B), and oxacillinases (OXA, Class D) are the major types. These enzymes confer carbapenem resistance through hydrolysis.6-11 Giri S. et al12. identified carbapenemase genes in 98% of the CRE isolates. Interestingly, most Klebsiella pneumoniae isolates possess dual carbapenemase genes (blaNDM and blaOXA 48).12 Bakthavatchalam et al1 identified OXA-48 in 52% of K. pneumoniae isolates, while dual OXA-48 and NDM genes were identified in 27% of the isolates, whereas in case of Escherichia coli isolates 68% NDM and 24% OXA-48 were identified.1 The focus of this study was to compare the accuracy and efficiency between different phenotypic methods such as the modified Hodge test (MHT), Carba NP test (strip test and Eppendorf tube test), and modified carbapenem inactivation method (mCIM) in the detection of carbapenemase enzyme production in Enterobacterales and to compare the results with the Kirby Bauer disc diffusion method for identification of carbapenem resistance.
This prospective study was conducted at the SRM Medical College Hospital and Research Centre, Tamil Nadu. A total of 150 Enterobacterales spp. isolated from 230 consecutive clinical specimens collected at the laboratory between April and May 2021 were inspected. Non-Enterobacterales bacteria isolated from the cultures were excluded from the study. The identification of the Enterobacterales isolates was performed by phenotypic confirmation using colony morphology on 5% sheep blood agar, MacConkey agar, as well as by manual biochemical tests. Patient demographics were collected from the laboratory database for analysis. No specific patient identifiers were used. Clinical samples including urine, pus/wound swabs, sputum, ear swabs, tracheal aspirates, tissue, and vaginal swabs, which resulted in the growth of organisms belonging to the family Enterobacterales, were included in the study. Antimicrobial susceptibility testing was performed for all isolates using the Kirby-Bauer disk diffusion method according to the CLSI 2021 guidelines. Phenotypic methods for carbapenemase detection were performed using the modified Hodge test (MHT), modified carbapenem inactivation method (mCIM), and the Carba NP test. The sensitivity and specificity of the phenotypic tests were analyzed using the Kirby Bauer disc diffusion method, using the CLSI 2021 guidelines as the gold standard for the detection of carbapenem resistance.
Performance of the modified Hodge test (MHT)
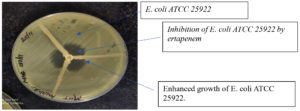
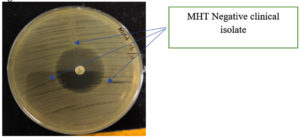
A 1:10 dilution of 0.5 McFarland E. coli (ATCC 25922) was cultured on Mueller-Hinton agar (MHA) plates using a sterile cotton swab. A 10 μg ertapenem disk was placed in the center of the MHA plate. Clinically confirmed Enterobacterales test isolates were inoculated from the edge of the disk to the edge of the plate. Three isolates were streaked onto an MHA plate. The plate was incubated overnight at 37°C, and the presence of clover leaf-like indentation of E. coli-susceptible strains growing along the streak of the test organism within the disk diffusion zone of ertapenem was considered positive (Figure 1). The absence of E. coli growth along the streak of the test organism within the disc diffusion zone was considered negative13 (Figure 2). This test is a modified version of the original Hodge test, which utilizes 10 μg imipenem disks. The original Hodge test was less sensitive in identifying carbapenemases compared to the modified Hodge test.14
Performance of the modified carbapenem inactivation method (mCIM)
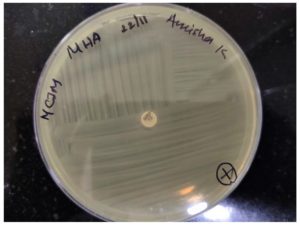
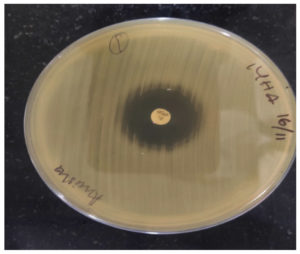
Following overnight incubation, 1 μL loopful of test bacteria was transferred to a tube containing 2 mL of trypticase soy broth (TSB) and the suspension was vortexed. A 10 μg meropenem disk was added to the suspension. The TSB-disk suspension was incubated for 4 h at 37°C. A 0.5 McFarland dilution of E. coli ATCC® 25922 was cultured on an MHA plate. The meropenem disk was removed from the TSB suspension and placed on an MHA plate inoculated with E. coli ATCC 25922. The plates were incubated overnight at 37°C. The presence of a zone diameter of 6–15 mm was considered positive (Figure 3) and a zone diameter of ≥19 mm was considered negative (Figure 4). This method is a modification of the carbapenem inactivation method described by Pierce et al15. The advantages of the mCIM over the CIM test include its high sensitivity (93%) and specificity (100%), compared to the CIM test, which has 82% sensitivity and 100% specificity. In addition, mCIM showed 92% sensitivity in the detection of OXA-48 carbapenemase compared to 39% sensitivity with the CIM test.15
Performance of Carba NP Test16
The Carba NP test requires the preparation of several solutions. The following solutions were prepared for the Carba NP assay: (a) Solution A (Concentrated solution of 0.5% w/v phenol red with 0.1 mmol/L ZnSO4); for this solution, a concentrated solution of 0.5% w/v phenol red (0.5 g in 100 mL distilled water was prepared. Next, 2 ml of concentrated phenol red solution was mixed in 16.6 ml of distilled water. Drops of 1 N sodium hydroxide solution were used to adjust the pH to 7.8. Following this, 180 μL of 10 mM zinc sulfate was added to obtain a final concentration of 0.1 mM. (b) Solution B was prepared by adding 6 mg/mL of imipenem to solution A. (c) 20 mM Tris-HCl lysis buffer.
Carba NP tube method
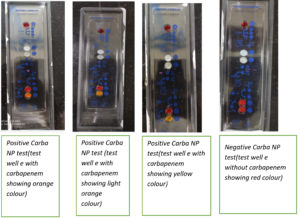
The bacterial colony grown overnight on MHA was scraped off using a loop, suspended in 100 μL of 20 mM Tris-HCl lysis buffer, and mixed for 5 s using a vortex. Next, 100 μL of Solution A was added to the first, and 100 μL of solution B was added to the second Eppendorf tube. Both tubes were then incubated at 37°C for 2 h. The development of red color in tubes A and B following incubation was considered negative. The development of red color in tube A and orange/yellow color in tube B following incubation was considered positive. The test cannot be interpreted when both tubes are yellow and was termed undetermined in such cases.
Carba NP strip method
This Strip method was performed using the Carba NP test kit on 20 Enterobacterales isolates. The RAPIDEC Carba NP test kit (Biomerieux, India) was used in this study. First, a rehydration step was performed. One isolate was tested on the Carba NP test strip. After removing the strip from the packaging, one ampule of API suspension was opened, and 100 µL was pipetted into wells ‘A’, ‘B’, and ‘C’ on the strip. The wells were covered by an incubation strip and incubated at 25°C for 4–10 min. The contents of well ‘B’ were stirred using a stick. Well ‘C’ was inoculated with the colonies to be tested. Colonies were added to well ‘C’ until the turbidity formed was similar to that of well ‘B’. The strip was incubated for 30 min at 25°C. Then, 25 µL was transferred from well ‘C’ to wells ‘D’ and ‘E’. A total of 25 µL was transferred from well ‘A’ to wells ‘D’ and ‘E’, respectively. The test strip was covered with the incubation lid and incubated at 37°C for 30 min. The development of a red color in wells ‘D’ and ‘E’ was considered negative. Red color in well ‘D’ and yellow in well ‘E’ was considered positive. The test cannot be interpreted when both wells ‘D’ and ‘E’ are yellow in color and such tests were termed undetermined (Figure 5).
A total of 150 Enterobacterales spp. isolated from 230 consecutive clinical specimens collected at the laboratory between April and May 2021 were included in the study. Non Enterobacterales isolates were excluded from the study. Enterobacterales were identified in various samples of patients belonging to different age groups. The majority of isolates were obtained from patients above 60 y (n=55; 37%), and from male patients (78; 52%). The sample distribution was as follows: urine (n=51; 34%), wound/pus (n=60; 40%), sputum/tracheal aspirate (n=18; 12%), vaginal swab (n=9; 6%), ear swab (n=8; 5.3%), tissue aspirate (n=2, 1.33%), and throat swab (n=2; 1.33%). Enterobacterales were isolated from various locations in the hospital of which majority of the isolates were from the general surgery ward (n=40; 26.6%) and intensive care units (ICU) (n=28; 18.66%). Among the Enterobacterales, organisms isolated included E. coli (n=45; 30%), K. pneumoniae (n=55; 36.6%), Enterobacter spp. (n=22; 14.6%), Citrobacter spp. (n=13, 8.6%) , Proteus mirabilis (n=10; 6.6%), Proteus vulgaris (n=3; 1.3%), and Morganella morganii (n=2; 1.3%).
Kirby Bauer disc diffusion test, according to CLSI 2021 guidelines, was performed on 150 Enterobacterales isolates to identify carbapenem resistance. Among the 150 isolates, 66 (44%) clinical samples were identified as CRE. Among the 66 CRE isolates, 35 (53%) were obtained from females and 31 (47%) from male patients. The distribution of CRE isolates was as follows E. coli (n=29; 44%), K. pneumoniae (n=36; 55%), Enterobacter (n=1, 1%). The highest incidence of CRE was observed in patients above 60 y (n=19; 28.78%). Of the 66 CRE isolates obtained, 11 were from intensive care units (17%) and 55 were from the general wards (83%). Distribution of the CRE isolates within the general wards were as follows: general medicine ward (n=8, 12.12%), general surgery ward n=14, 21.21 %), obstetrics and gynecology ward (n=8, 12.12%), orthopedics ward (n=2, 3.03%), nephrology ward (n=3, 4.54%), urology ward (n=13, 19.69%), pediatrics ward (n=2, 3.03%), ENT ward (n=3, 4.54%), pulmonology ward (n=2, 3.03%). The highest number of CRE isolated were obtained from the samples of patients from the general surgery ward, followed by the urology ward. The distribution of CRE isolates among the clinical samples was as follows: urine (n=20; 30.30%), wound/pus swab (n=27; 40.90%), sputum/tracheal aspirate (n=7; 10.60%), vaginal swab (n=7, 10.60%), ear swabs (n=2; 3.03%), tissue aspirate (n=1; 1.51%), throat swabs (n=2; 3.03%).
Carbapenemase enzyme was detected using the MHT, mCIM, and Carba NP tests. Of the 150 Enterobacterales isolates, 63 (42%) were MHT positive and 87 (54%) were MHT negative. The distribution of positive MHT isolates was as follows: E. coli (n=28; 44.44%), K. pneumoniae (n=34; 53.96%), and Enterobacter spp. (n=1; 1.58%). The mCIM test was positive in 64 (42.66%) and negative in 86 (57.33%) isolates. The distribution of positive mCIM isolates was as follows: E. coli (n=30; 46.87%), K. pneumoniae (n=33; 51.56%), and Enterobacter spp. (n=1; 1.56%). A total of 11 CRE isolates were positive in the Carba NP strip test, which was performed on 20 isolates, and 54 isolates were positive in the Carba NP tube test, which was performed on 130 isolates. In total, both the methods of Carba NP test identified 65 (43.33%) CRE isolates, amongst which E. coli (n=30; 46.15%), K. pneumoniae (n=34; 52.30%), Enterobacter spp. (n=1; 1.53%) were the predominant bacteria. In summary, of the total isolates, 66/150 (44%) CRE isolates were detected using the Kirby Bauer disc diffusion test, 62/150 (41%) using MHT, 64/150 (43%) using mCIM, and 65/150 (43%) using Carba NP test (Table). Kirby–Bauer disc diffusion testing was used as the gold standard for detecting carbapenem resistance. The sensitivity and specificity of MHT test, mCIM, and Carba NP within 95% CI were 93.94%/100%, 96.97%/100%, and 98.48%/100%, respectively. The positive and negative predictive values of MHT test, mCIM, and Carba NP were 100%/95.45%, 100%/97.67%, and 100%/98.82% respectively. The accuracies of the MHT, mCIM, and Carba NP test were 97.33%, 98.67%, and 99.33% respectively. The measure of agreement (kappa value) between disc diffusion and Carba NP, mCIM, and MHT were 0.986, 0.973, and 0.946, respectively, indicating good agreement between the tests.
Table :
Comparison Of Disc Difussion Testing With Modified Hodge Test (MHT), Modified Carbapenem Inactivation Method (mCIM), Carba NP Test For Detection Of Carbapenem Resistant Enterobacterales (N=150).
| Result | MHT | mCIM | CARBA NP | ||||
|---|---|---|---|---|---|---|---|
| +VE | -VE | +VE | -VE | +VE | -VE | ||
| Disc Diffusion | +Ve | 62 | 4 | 64 | 2 | 65 | 1 |
| -Ve | 0 | 84 | 0 | 84 | 0 | 84 | |
Carbapenems, such as ertapenem, imipenem, and meropenem, are β-lactam antibiotics used for the treatment of multidrug-resistant bacterial infections (MDR). Carbapenem resistance in gram-negative bacteria poses a considerable threat to human health. The misuse of antibiotics in healthcare and agriculture has been a major factor contributing to the development of carbapenem resistance across the world.17
Carbapenemase detection is important in the identification of carbapenemase-producing CRE (CP-CRE) and non-carbapenemase-producing CRE (non-CR-CRE). Tamma et al18 compared the outcomes of patients with CP-CRE and non-CP-CRE bacteremia and identified that the odds of mortality within 14 days were four fold higher in CP-CRE than that in non-CP-CRE bacteremic patients.18
A steady increase in the prevalence of CRE has been reported in centers across India. Datta et al19 reported a CRE prevalence of 7.87% in North India in the year 201219; Nair PK et al20 reported a CRE prevalence of 12.26% in Western India in 2013.20 Jan R et al21. reported 8% CRE prevalence in southern India in 2016.21 Rao A and Indumathi et al22 reported 13.95% CRE prevalence in Southern India in 2016.22 Khare V et al23 reported 37.9% CRE prevalence in northern India in 2017.23 Pawar SK et al24 reported 31.77% CRE prevalence in western India in 2018.24 This study showed a CRE prevalence of 44% in our center, and the above data point towards a rapid increase in the prevalence of CRE strains across India. CRE has been frequently isolated from patients admitted to general wards. In the present study, proportions of CRE obtained from intensive care units was 11/66 (17%) and that from general wards were 55/66 (83%), which was similar to that identified by Nair et al20 in 2013, wherein they detected CRE in 26% of samples from intensive care units and 42% from general ward patients.20
Of the different tissues used for sample collection, majority of CRE were isolated from wound/ pus swabs (40.90%) followed by urine samples (30.30%) in this study. Samples were collected from patients belonging to five different age groups. The highest number of CRE isolates were obtained from the patients above 60 years (28.78%), followed by the 21-30 y (19.69%) and 31–40 y (18.18%) groups, unlike previous studies, indicating a rise in resistance in the older individuals. For example, Patidar et al25 2021, reported the highest prevalence of CRE in the age group of 21-40 y (36%). In the present study, K. pneumoniae (55%) was the most predominant isolate among the CRE pathogens. Urine (41%), followed by endotracheal aspirate (25%), yielded the highest number of CRE isolates. Carbapenem resistance was the highest in Enterobacter spp.(40%) in this study.25 Nair et al20, also reported the highest number of CRE in urine specimens (46%).20 Thomas et al26, identified 52.1% CRE in urine specimens, and E. coli (63.75%) was the predominant isolate in this study. The study also reported a higher percentage of CRE in the age group of 21-40 y (36.25%).26 Modi CM et al27, reported 29% CRE from urine samples, with Klebsiella spp. (51%) as the predominant isolate.27
MHT, mCIM, and Carba NP tests were performed to identify carbapenemase production from 150 Enterobacterales isolates in this study. MHT was positive in 62/150 (41%) isolates, mCIM was positive in 64/150 (43%), and Carba NP was positive in 65/150 (43%) isolates. According to the CLSI 2021 guidelines, disc diffusion testing for carbapenems using the current breakpoints identifies CRE and does not require laboratories to perform routine phenotypic tests for the detection of carbapenemases.16 In the present study, the sensitivity and specificity of the MHT test, mCIM, and Carba NP within 95% CI were 93.94%/100%, 96.97%/100%, 98.48%/100%, respectively. Bartolini et al28, reported 94% sensitivity and 100% specificity for MHT, similar to the present study.28 A pilot study on carbapenemase detection conducted by Pragasam et al29, found the specificity and sensitivity of Carba NP to be 100% and 81–94% for CP-CRE.29 Kumudunie et al30, identified the sensitivity and specificity of MHT, mCIM, and Carba NP tests to be 90.7%/92.1%, 100%/100%, and 75.9%/100%, respectively. This study documented that the mCIM test is the most sensitive test for the detection of CRE.30 Beig et al31 documented Carba NP and mCIM tests as highly sensitive and specific phenotypic tests for detection of CRE with 97% and 94% sensitivity compared to MHT with 67% sensitivity.31 However, in the present study, both mCIM and Carba NP were highly sensitive in the detection of CRE.
Genotypic tests are considered the gold standard for carbapenemase detection; however, as they are relatively expensive, they are difficult to set up in routine clinical laboratories. Although previous studies had reported challenges in the interpretation of MHT and have expressed concerns regarding false positivity of the test,32-34 the present study had zero false-positive results for MHT. The limitation of the MHT and mCIM tests, however, is that they are time consuming, as interpretation of results is performed after overnight incubation (18–24 hours), unlike Carba NP test, which can be read after two hours of incubation.
Molecular techniques are considered as gold standards for the identification of CRE; however, owing to financial constraints, most laboratories fail to set them up and use them routinely. In such cases, phenotypic detection methods for carbapenemases, which possess high sensitivity and specificity rates, help in the rapid and cost-effective identification of CRE. Among the phenotypic tests for carbapenemase detection, the mCIM and Carba NP tests showed higher sensitivity in detecting CRE compared to MHT.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
All authors have made a substantial, direct and intellectual contribution to the work and approved it for publication
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during the study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, SRM Medical College Hospital and Research Centre, Tamil Nadu, India with wide reference number 299/IEC/2021.
- Bakthavatchalam YD, Routray A, Mane A, et al. In vitro activity of Ceftazidime-Avibactam and its comparators against Carbapenem resistant Enterobacterales collected across India: results from ATLAS surveillance 2018 to 2019. Diagn Microbiol Infect Dis. 2022;103(1):115652.
Crossref - Nordmann P, Naas T, Poirel L. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-1798.
Crossref - Birgy A, Bidet P, Genel N, et al. Phenotypic screening of carbapenemases and associated β-lactamases in carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2012;50(4):1295-1302.
Crossref - Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae: a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873-5884.
Crossref - Guilarde AO, Turchi MD, Martelli CMT, Primo MGB, Batista LJA. Bacteremias em pacientes internados em hospital universitario. Rev Assoc Med Bras. 2007;53(1):34-38.
Crossref - Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289(1036):321-331.
Crossref - Ambler RP, Coulson AF, Frere JM, et al. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276(Pt 1):269-270.
Crossref - Dienstmann R, Picoli SU, Meyer G, Schenkel T, Steyer J. Avaliacao fenotipica da enzima Klebsiella pneumoniae carbapenemase (KPC) em Enterobacteriaceae de ambiente hospitalar. J Bras Patol Med Lab. 2010;46(1):23-27.
Crossref - Singh M, Kakati B, Agarwal RK, Kotwal A. Detection of Klebsiella pneumoniae carbapenemases (KPCs) among ESBL/MBL producing clinical isolates of Klebsiella pneumoniae. Int J Curr Microbiol App Sci. 2015;4(4):726-731.
- Shields RK, Potoski BA, Haidar G, et al. Clinical outcomes, drug toxicity and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis. 2016 2;63(12):1615-1618.
Crossref - Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2014;20(9):862-867
Crossref - Giri S, Karade S, Sen S. Genotypic characterization of carbapenem resistant Enterobacterales in clinical isolates from western Maharashtra. Indian J Med Microbiol. 2021;39(4):500-503.
Crossref - Amjad A, Mirza Ia, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3(4):189-193.
- Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003;41(10):4623-4629.
Crossref - Pierce VM, Simner PJ, Lonsway DR, et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J Clin Microbiol. 2017 ;55(8):2321-2333.
Crossref - CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2021.
- Watkins RR, Bonomo RA. Overview: Global and Local Impact of Antibiotic Resistance. Infect Dis Clin North Am. 2016;30(2):313-322.
Crossref - Tamma PD, Goodman KE, Harris AD, et al. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin Infect Dis. 2017;64(3):257-264.
Crossref - Datta P, Gupta V, Garg S, Chander J. Phenotypic method for differentiation of carbapenemases in Enterobacteriaceae: study from north India. Indian J Pathol Microbiol. 2012;55(3):357-360.
Crossref - Nair PK, Vaz MS. Prevalence of carbapenem resistant Enterobacteriaceae from a tertiary care hospital in Mumbai, India. J Microbil Infect Dis. 2013;3(04):207-210.
Crossref - Jan R, George N, Mathew M, et al. Prevalence of carbapenem-resistant Enterobacteriaceae in a tertiary care referral centre: Kerela, South India. Int J Curr Res. 2016;8(12):44353-44355.
- Rao A, Indumathi VA.Detection of carbapenem resistant Enterobacteriaceae from clinical isolates. Int J Curr Microbiol App Sci. 2016;5(5):864-869.
Crossref - Khare V, Gupta P, Haider F, Begum R. Study on MICs of Tigecycline in Clinical Isolates of Carbapenem Resistant Enterobacteriaceae (CRE) at a Tertiary Care Centre in North India. J Clin Diagn Res. 2017;11(3):DC18-DC21.
Crossref - Pawar SK, Mohite ST, Shinde RV, Patil SR, Karande GS. Carbapenem – resistant Enterobacteriaceae: Prevalence and bacteriological profile in a tertiary care teaching hospital from rural western India. Ind J Microbiol Res. 2018;5(3):342-347.
Crossref - Patidar N, Vyas N, Sharma S, Sharma B. Phenotypic Detection of Carbapenemase Production in Carbapenem-Resistant Enterobacteriaceae by Modified Hodge Test and Modified Strip Carba NP Test. J Lab Physicians. 2021;13(1):14-21.
Crossref - Thomas N, Sarwat T. Prevalence of carbapenem resistant Enterobacteriaceae in a tertiary care hospital. Int J Curr Microbiol App Sci. 2019;8(11):1418-1424.
Crossref - Modi CM, Singh SP, Pandya YG, Patel CP, Patel RM. Prevalence of Carbapenem Resistant Enterobacteriaceae in a Tertiary Care Hospital of Gujarat, India. J Clin of Diagn Res. 2021;15(3):DC11-DC14.
Crossref - Bartolini A, Frasson I, Cavallaro A, Richter SN, Palu G. Comparison of phenotypic methods for the detection of carbapenem non-susceptible Enterobacteriaceae. Gut Pathog. 2014;6:13.
Crossref - Pragasam AK, Sahni RD, Anandan S, et al. A Pilot Study on Carbapenemase Detection: Do We See the Same Level of Agreement as with the CLSI Observations. J Clin Diagn Res. 2016;10(7):DC09-DC13.
Crossref - Kumudunie WGM, Wijesooriya LI, Wijayasinghe YS. Comparison of four low-cost carbapenemase detection tests and a proposal of an algorithm for early detection of carbapenemase-producing Enterobacteriaceae in resource-limited settings. PLoS One. 2021;16(1):e0245290.
Crossref - Beig M, Taheri M, Arabestani MR. Comparison of Different Phenotypic Tests versus PCR in the Detection of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Hamadan, Iran. Int J Microbiol. 2021;2021:5582615.
Crossref - Carvalhaes CG, Picao RC, Nicoletti AG, Xavier DE, Gales AC. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65(2):249-251
Crossref - Pournaras S, Poulou A, Tsakris A. Inhibitor-based methods for the detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice by using boronic acid compounds. J Antimicrob Chemother. 2010;65(7):1319-1321.
Crossref - Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(2):477-479.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.